Abstract
Aims/hypothesis
We hypothesised that maternal diabetes impairs kidney formation in offspring via augmented expression of hedgehog interacting protein (HHIP). Our gene-array results were performed in neonatal kidneys from our murine model of maternal diabetes and indicated that Hhip expression was significantly modulated by maternal diabetes.
Methods
We systematically examined the functional role of HHIP in kidney formation in our murine maternal diabetes model and elucidated the potential mechanisms related to dysnephrogenesis in vitro.
Results
The kidneys of the offspring of diabetic dams, compared with those of the offspring of control non-diabetic dams, showed retardation of development—small kidneys and less ureteric bud (UB) branching morphogenesis. Augmented HHIP expression was observed in the offspring of diabetic dams, initially localised to differentiated metanephric mesenchyme and UB epithelium and subsequently in maturing glomerular endothelial and tubulointerstitial cells. The heightened HHIP targeting TGF-β1 signalling was associated with dysmorphogenesis. In vitro, HHIP overexpression decreased sonic hedgehog and paired box gene 2 proteins (SHH and PAX2, respectively) and increased transcriptional nuclear factor-kappa B (NFκB, p50/p65), phosphorylation of p53, and TGF-β1 expression. In contrast, overexpression of PAX2 inhibited HHIP and NFκB and activated SHH, N-myc and p27Kip1 expression. Moreover, high glucose stimulated HHIP expression, and then targeted TGF-β1 signalling. Thus, PAX2, via a negative autocrine feedback mechanism, attenuated the stimulatory effect of high glucose on HHIP expression.
Conclusions/interpretation
Maternal diabetes modulates kidney formation in young progeny mediated, at least in part, via augmented HHIP expression.
Similar content being viewed by others
Introduction
Renal malformations are responsible for 34–59% of chronic kidney disease in children and for 31% of paediatric end-stage renal disease in the USA [1–3]. Of the many factors associated with congenital abnormalities of the kidney and urinary tract, diabetes mellitus during gestation seems to be one of the key players in both human and experimental settings [4]. For example, when the normal pattern of kidney formation is interrupted in both humans and experimental animal models (e.g. by a high ambient in utero glucose level), kidney abnormalities with a low total nephron number may ensue, either in isolation or as part of multiple malformation syndromes. The incidence of such anomalies appears to be proportional to the degree of maternal hyperglycaemia [5–7]. The underlying mechanisms of such findings are incompletely delineated [6, 8, 9].
We previously reported that severe maternal diabetes (maternal blood glucose concentration >30 mmol/l) in a murine model (C57/BL6 background) impairs kidney formation and is associated with an intrauterine growth restriction (IUGR) phenotype in the offspring. Affected pups have relatively small kidneys/body weight with ~ 40% fewer nephrons [6, 10, 11]. We performed screening gene profiling of IUGR neonatal kidneys from offspring of dams with diabetes vs kidneys from offspring of control non-diabetic dams (Affymetrix GeneChip Mouse Gene 1.0ST array platform, Affymetrix, Santa Clara, CA, USA), aiming to identify the novel proteins or signalling pathways through which maternal diabetes impairs nephrogenesis.
We analysed our ontology database data using the following: the Affymetrix Expression Console (www.affymetrix.com) to normalise the gene database; the Robust Multi-Array Analysis (RMA) algorithm (Partek, St. Louis, Missouri, USA, www.partek.com) to compute expression values; a TM4 MultiArrayViewer (Boston, MA, USA, www.tm4.org) to analyse differential expression (p value threshold of 0.01 and false discovery rate of 0.05). Of the genes showing significant differences between the groups in our murine maternal diabetes model, Hhip (fold change, 1,536; p = 0.007) was upregulated in the neonatal kidneys of offspring from diabetic dams but was expressed normally in insulin-treated dams, as it is highly related to development, signal transduction and proliferation in the analysis of functional pathway clustering.
Hhip, which encodes hedgehog interacting protein (HHIP), a transmembrane glycoprotein, was discovered by screening a mouse cDNA expression library for proteins that bound to sonic hedgehog protein (SHH) [12]. As a putative antagonist of the hedgehog signal pathway, HHIP binds to proteins from three mammalian hedgehogs—SHH, Indian hedgehog (IHH) protein and desert hedgehog (DHH) protein—to attenuate their bioactivity [12]. Of the three mammalian hedgehog genes, only expression of Shh and Ihh has been reported in the developing murine kidney. However, the absence of a renal phenotype in Ihh-deficient mice indicates that it might not have a key role in kidney development [13]. SHH signalling optimally controls the expression of three distinct classes of genes expressed in the nephrogenic zone that are required for normal renal development: early kidney inductive and patterning genes (Pax2, Sall1); cell-cycle modulators (Ccnd1, Mycn) and the signalling effectors of the hedgehog pathway (Ci homologues, Gli1, Gli2) [14].
The protein encoded by Hhip is expressed in vascular endothelial cells [15] and in cells adjacent to those expressing SHH, positioning it to bind to the SHH [12]. To date, although evidence indicates that cellular expression of HHIP inhibits the activation of SHH signalling and promotes angiogenesis during the formation of a variety of tumours [15, 16], the functional role of Hhip expression in developing and mature kidney is incompletely understood.
In the present studies, we first validated the changes in HHIP expression in pregnancies complicated with diabetes and then investigated whether ectopic Hhip gene upregulation induced by maternal diabetes could interfere with SHH-targeting gene signalling [14]. We also investigated whether upregulation of HHIP had an impact on kidney formation from birth to 3 weeks in young offspring since in rodents nephrogenesis continues until postnatal day 10 (i.e. murine gestational period between 19 and 21 days) [5]. We then examined potentially involved mechanisms using both in vivo and in vitro studies.
Methods
Animal models
We used the Hoxb7–green fluorescent protein (GFP) transgenic (Tg) (Hoxb7-GFP-Tg, C57/BL6 background) mouse for the current study; this mouse is fertile with a normal phenotype at birth and during adult life [6, 7]. Hoxb7-GFP-Tg mice (GFP expression specifically in ureteric bud [UB] driven by Hoxb7 promoter), provided by F. Costantini (Columbia University Medical Center, New York, NY, USA) [17, 18], were engineered to allow UB branching morphogenesis to be visualised in real time in vivo as reported previously [6, 7].
As reported previously, we used a murine maternal diabetes model [6, 7, 11] induced by a single intraperitoneal injection of streptozotocin (STZ) (Sigma-Aldrich Canada, Oakville, ON, Canada) at a dose of 150 mg/kg of body weight at E13 gestation age. Neonatal kidneys of offspring from three groups of dams (non-diabetic, diabetic and diabetic dams treated by insulin implantation at E15 [Linshin, Scarborough, ON, Canada]) were used.
Animal care and the procedures used were approved by the Institutional Animal Care Committee of the CRCHUM. Mice were housed under standard humidity and lighting conditions (12 h light–dark cycles) with free access to standard mouse chow and water.
Cell lines
The MK4 cell line [11, 19] was obtained from S. S. Potter (Children’s Hospital Medical Center, Cincinnati, OH, USA) and is representative of late embryonic metanephric mesenchyme (MM), as it undergoes mesenchymal-to-epithelial conversion. MK4 cells express genes typical of late MM, including Pax2, Pax8, Wnt4, Cdh6, collagen IV and Lfb3 (also known as Hnf1) [20]. Mouse UB tip cells were provided by J. Barasch (Columbia University, New York, NY, USA) and were generated from microdissected UBs of E11.5 mouse embryos transgenic for simian virus 40 large T antigen (Immorto-mouse) and which express epithelial and ureteric genes (Ret, Met) without hepatocyte growth factor [21]. The plasmids pcDNA3.1/Hhip and pcDNA3.1/Pax2 were gifts from A. McMahon (Harvard University, Cambridge, MA, USA) [12] and P. Goodyer (McGill University, Montreal, QC, Canada) [22], respectively.
We used both MK4 and UB tip cells to represent the MM and UB epithelium, respectively, and examined Hhip gene expression and HHIP regulation with or without a high glucose (25 mmol/l d-glucose) insult in MK4 cells. Unless otherwise indicated, all control cells were incubated in low-glucose (5 mmol/l d-glucose) medium containing 20 mmol/l d-mannitol to ensure equi-osmolar control.
Counting of UB tips and measurement of neonatal kidney size
Neonatal kidneys of offspring from three groups of dams (non-diabetic, diabetic and diabetic dams treated by implanted insulin) were dissected aseptically and the number of UB tips in each group was assessed as reported previously [7]. Neonatal kidney size (length [mm], width [mm] and area [mm2]) was measured from the two-dimensional GFP images of freshly isolated neonatal kidneys using Image-J 1.48 (NIH, USA, http://imagej.nih.gov/ij/).
Real-time quantitative PCR
Real-time quantitative PCR (qPCR) (the Fast SYBR green mastermix kit and the 7500 Fast real-time PCR system; Applied Biosystems, Life Technologies, Foster City, CA, USA) was performed as reported previously [6, 7, 11]. The final real-time qPCR product was also reviewed by Typhoon Trio Variable Mode Imager (GE Health Care, Bio-Sciences, Uppsala, Sweden). The following primers were used: Hhip forward 5′-CCCATCGGCTCTTCATTCTA-3′, reverse 5′-CCTTTCGTCTCCTCCCTTTA-3′ (NM_020259.4).
Western blot, immunohistochemistry and immunofluorescence
Western blot analysis, immunohistochemical staining and immunofluorescence were performed essentially according to an established procedure as reported previously [6, 7, 11, 19]. The following antibodies were used: monoclonal anti-HHIP and β-Actin antibodies (Sigma-Aldrich Canada); Dolichos biflorus agglutinin (DBA)–FITC (Vector Laboratories, Burlington, ON, Canada) staining for UB identification [7]; anti-SHH, -nuclear factor-kappa B (NFκB) (p50/p65), -N-myc, -CD31 and -p53 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-paired box gene 2 protein (PAX2) antibody (Covance, Richmond, CA, USA); anti-phospho-p53 (Ser 15) (Abcam, Cambridge, MA, USA); anti-p27Kip1 (BD Transduction Laboratories, Mississauga, ON, Canada); anti-TGF-β1 antibody (R&D Systems, Burlington, ON, Canada); phospho-Smad2 (Ser465/467)/Smad3 (Ser423/425) antibody (New England Biolabs, Whitby, ON, Canada) and Smad2/3 antibody (Cedarlane-Millipore, Burlington, ON, Canada).
Statistical analysis
Statistical significance between the experimental groups was analysed by one-way ANOVA, followed by the Bonferroni test, using Graphpad Software, Prism 5.0 (www.graphpad.com/prism/Prism.htm). Three or four separate experiments were performed for each protocol. Data are expressed as means ± SD. A probability level of p ≤ 0.05 was considered to be statistically significant.
Results
UB branching morphogenesis
We compared UB branching morphogenesis in neonatal kidneys of three subgroups of Hoxb7-GFP-Tg murine lines’ offspring (Con-offspring, the offspring of non-diabetic dams; Dia-offspring, the offspring of diabetic dams; Ins-offspring, the offspring of diabetic dams treated by insulin implantation) (Fig. 1a). At birth, maternal blood glucose concentration (mmol/l) was as follows: non-diabetic dams, 9.03 ± 1.75; diabetic dams, 29.36 ± 2.54; diabetic dams treated by insulin implantation, 15.8 ± 1.36.
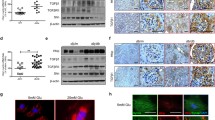
(a) GFP images of freshly isolated neonatal kidney (magnification ×4). (b) Neonatal kidney size; Length, L; Width, W; Area, A. (c) The number of UB tips in neonatal kidney. Circles, Con-offspring; squares, Dia-offspring; inverted triangles, Ins-offspring. (d, e) Real-time qPCR analysis of Hhip mRNA expression in neonatal kidney. (d) Typhoon gel image. (e) The relative density of Hhip mRNA was compared with its own β-actin mRNA. Con-offspring values were considered as 100%. Each point represents the mean ± SD of three independent experiments. **p ≤ 0.01; ***p ≤ 0.001
Compared with Con-offspring, Dia-offspring with IUGR phenotype had smaller neonatal kidney/body weight ratio as previously reported [6], smaller kidney size (Fig. 1b) and fewer UB tips (Con-offspring, 859.8 ± 7.74; Dia-offspring, 669.9 ± 18.96) (Fig. 1c). The impairment of UB branching was ameliorated by insulin administration to the mothers (Ins-offspring, 807.7 ± 20.52) (Fig. 1c).
Hhip gene expression and localisation in vivo
We subsequently validated and confirmed Hhip mRNA expression by real-time qPCR (Fig. 1d, e) and HHIP protein expression by immunohistochemistry (Fig. 2a) and immunofluorescence staining (Fig. 2b). Consistent with the data obtained from our prior microarray analysis, HHIP expression was significantly increased in the neonatal kidneys of IUGR Dia-offspring.
(a) Immunohistochemical staining showing HHIP expression in neonatal kidneys. Black frame, Con-offspring; red frame, Dia-offspring; blue frame, Ins-offspring (magnification ×4, ×10 and ×60, as shown). (b, c) Co-localisation, by immunofluorescence, of HHIP (red arrows) and DBA (green arrows) as well as HHIP/DBA merge in UB tips in renal cortex (b) and UB trunks in renal medulla (c) of the neonatal kidneys of Dia-offspring (magnification ×60)
We next followed the time course of intrarenal HHIP expression and its localisation pattern during late kidney formation in the offspring (from day 1 to 3 weeks). In the kidneys of Con-offspring, HHIP expression was detected in both UB and MM lineages on day 1 (Fig. 2) and in the collecting duct system at 1 week (Fig. 3). After kidney formation was completed (3 weeks), very limited HHIP expression was observed and was localised to mature glomerular endothelial and adjacent tubulointerstitial cells (Fig. 4).
HHIP expression in 3-week-old kidneys. (a) Immunohistochemical staining showing HHIP. Black frame, Con-offspring; red frame, Dia-offspring; blue frame, Ins-offspring (magnification ×20 and ×60). (b) Co-localisation, by immunofluorescence, of HHIP (red) and CD31 (green), as well as HHIP/CD31 merge (yellow), in 3-week-old kidneys of Dia-offspring (magnification ×60)
In contrast, the enhanced HHIP expression remained in the nephrogenic zone and was detected in both UB and MM epithelium in the kidneys of Dia-offspring (Fig. 2a). An increased amount of secreted HHIP and intracellular HHIP was detected adjacent to UB tips (Fig. 2b) and UB trunks (Fig. 2c) (DBA–FITC staining for UB identification [7]) in the neonatal kidneys. By 1 week of age, this increased HHIP expression was progressively evident in glomeruli, ultimately forming a nodular expression pattern (Fig. 3). At 3 weeks, the Dia-offspring had only small amounts of HHIP in the tubulointerstitium (Fig. 4a) but HHIP was strongly expressed in glomerular endothelial cells in the neonatal kidney cryosections, as assessed by immunofluorescence (confirmed by co-localisation with CD31, a marker of endothelial cells) (Fig. 4b). Furthermore, the pattern of HHIP expression in the kidneys of Ins-offspring was essentially the same as that in the Con-offspring.
HHIP expression and regulation in vitro
Western blotting showed that HHIP, SHH and PAX2 were basally expressed in both MK4 and UB cells as well as in cells transfected with the empty vector plasmid, pcDNA3.1 (1 μg/ml) (Fig. 5a), a control for the rest of transfection experiments. Overexpression of Hhip by transient transfection of pcDNA3.1/Hhip (1 μg/ml) into both MK4 and UB tip cells (Fig. 5b) inhibited both SHH and PAX2 expression. Conversely, overexpression of PAX2 by transient transfection with pcDNA3.1/Pax2 (1 μg/ml) increased SHH and decreased HHIP expression (Fig. 5b).
HHIP, PAX2 and SHH protein expression analysed by western blot. (a) Basal expression in naive MK4 and UB cells as well as the cells transfected with pcDNA3.1 plasmid (1 μg/ml). (b) MK4 and UB tip cells were transfected with the plasmids pcDNA3.1 (white bars), pcDNA3.1/Hhip (black bars) and pcDNA 3.1/Pax2 (grey bars) at 1 μg/ml, respectively; the gene expression protein ratio in cells transfected with pcDNA3.1 plasmid was considered to be 100%. (c, d) The time course of high glucose effect on HHIP and SHH expression in naive MK4 cells (c) and UB tip cells (d) was analysed by western blot using 20 mmol/l d-mannitol (d-Man) as an osmotic control (white bars, 5 mmol/l d-glucose [d-Glu]/HHIP; black bars, 25 mmol/l d-glucose/HHIP; light-grey bars, 5 mmol/l d-glucose/SHH; dark-grey bars, 25 mmol/l d-glucose/SHH). The gene expression protein ratio in 5 mmol/l glucose medium at the 6 h incubation period were considered to be 100%. Each point represents the mean ± SD of three independent experiments. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
We tested the effect of a high-glucose milieu (25 mmol/l d-glucose) on HHIP and SHH expression in MK4 (Fig. 5c) and UB tip cells (Fig. 5d). High glucose persistently stimulated HHIP expression in a time-dependent manner, whereas SHH expression was only initially increased by high glucose within 24 h, then decreased rapidly after 48 h in both cell lines (Fig. 5c, d).
We then performed studies using the MK4 cell line, as it is derived from MM lineages, to examine high-glucose milieu regulation on HHIP expression, since we had noted that the augmented HHIP expression in the offspring of dams with diabetes was seen in the developing proximal portion of the nephron, which is mainly derived from differentiated MM lineages. High glucose (25 mmol/l d-glucose) stimulated HHIP expression in a time- and dose-dependent manner in naive MK4 cells but other glucose analogues such as l-glucose, d-mannitol and 2-deoxy-d-glucose did not (Fig. 6a–c). We also observed that insulin (10−6 mol/l) inhibited the stimulatory effect of high glucose on Hhip gene expression (as assessed by western blotting of HHIP) in MK4 cells (Fig. 6d).
Effects of high d-glucose (25 mmol/l, d-Glu) on HHIP expression in naive MK4 cells. (a) Time course analysed by western blot using 20 mmol/l l-glucose (l-Glu) as an osmotic control. (b) Effect of the glucose analogues d-glucose (d-Glu), l-glucose (l-Glu), d-mannitol (d-Man) and 2-deoxy-d-glucose (2-Deoxy), on HHIP expression analysed by western blot. (c) Dose-dependent effect of glucose analysed by western blot. (d) Insulin’s inhibitory effect analysed by western blot. The gene expression protein ratio in 5 mmol/l glucose medium with 20 mmol/l d-mannitol at the 24 h incubation period was considered to be 100%. Each point represents the mean ± SD of three independent experiments. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
We used MK4 cells transfected with the plasmid, pcDNA3.1 (MK4-vector) (1 μg/ml) as controls, since the basal expression in naive and transfected MK4 cells were essentially identical (Fig. 5). A high-glucose milieu stimulated transcriptional NFκB (p50/p65) and phosphorylation of p53 in MK4-vector cells. Overexpression of Hhip in MK4 cells alone significantly increased NFκB (p50/p65) and p53 expression, including phosphorylation of p53 (Fig. 7a), and this activation was further enhanced by exposure to high glucose. In contrast, overexpression of Pax2 in MK4 cells significantly decreased HHIP and NFκB (p50/p65) expression; further, this inhibition was unaffected by high glucose (Fig. 7b). In addition, overexpression of Pax2 in MK4 cells resulted in significantly increased SHH, N-Myc and p27kip1 expression and high-glucose exposure had no additional effect.
Effect of high glucose on different genes of MK4 transfected cells analysed by western blot. (a) MK4 cells transfected with the plasmids pcDNA3.1 (1 μg/ml) (MK4-Vector) in d-glucose (d-Glu) 5 mmol/l (white bars) and 25 mmol/l glucose (black bars) or pcDNA3.1/Hhip (1 μg/ml) (MK4-Hhip) in 5 mmol/l glucose (light-grey bars) and 25 mmol/l glucose (dark-grey bars). (b) MK4-Vector in 5 mmol/l glucose (white bars) and 25 mmol/l glucose (black bars); MK4 cells transfected with the plasmid pcDNA3.1/Pax2 (1 μg/ml) (MK4-Pax2) in 5 mmol/l glucose (light-grey bars) and 25 mmol/l glucose (dark-grey bars). The gene expression protein ratio in MK4-Vector cells in 5 mmol/l glucose medium with 20 mmol/l d-mannitol (d-Man) were considered to be 100%. Each point represents the mean ± SD of three independent experiments. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
TGF-β1 expression in vivo and in vitro
In vitro, we observed overexpression of Hhip in MK4 cells resulted in increased TGF-β1 and phosphorylated Smad2/3 expression and that this activation was further enhanced by high-glucose media (Fig. 8a). Recombinant TGF-β1 upregulated HHIP and phosphorylated Smad2/3, but downregulated p27Kip1 expression in a dose-dependent manner in naive MK4 cells (Fig. 8b). In vivo studies validated these findings—TGF-β1 expression was significantly increased in the kidneys of Dia-offspring from day 1 to 3 weeks of age (Fig. 8c). The heightened TGF-β1 expression displayed a similar immunohistochemical pattern to HHIP in mature glomerular endothelial and adjacent tubulointerstitial cells (Fig. 8d).
HHIP and TGF-β1 expression in vitro (a, b) and in vivo (c, d). (a) TGF-β1 expression in MK4 cells transfected with the plasmids pcDNA3.1 (1 μg/ml) (MK4-Vector) in 5 mmol/l glucose (white bars) and 25 mmol/l glucose (black bars) or pcDNA3.1/Hhip (1 μg/ml) (MK4-Hhip) in 5 mmol/l glucose (light-grey bars) and 25 mmol/l glucose (dark-grey bars). The gene expression protein ratio in MK4-Vector cells in 5 mmol/l glucose medium with 20 mmol/l d-mannitol (d-Man) was considered as 100%. (b) The dose–effect of recombinant TGF-β1 (rTGF-β1) on HHIP, phospho-Smad2/3, Smad 2/3 and p27Kip1 expression in naive MK4 cells. The gene expression protein ratio in rTGF-β1 (0 ng/ml) was considered as 100%. (c) Western blot performed in neonatal and 3-week-old kidneys of Con-offspring and Dia-offspring. Con-offspring values were considered as 100%. (d) Immunohistochemical analysis of TGF-β1 expression in 1-, 2- and 3-week-old kidneys of Con-offspring (black frame) and Dia-offspring (red frame) (magnification ×20). Each point represents the mean ± SD of three independent experiments. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001
Discussion
The present study suggests that heightened ectopic HHIP expression targets NFκB, p53 and TGF-β1 expression, which is associated with dysmorphogenesis in young Dia-offspring. We would suggest that overexpression of Pax2 may create a negative autocrine feedback on HHIP regulation and may attenuate the stimulatory effect of high glucose on Hhip expression in vitro.
Using the Hoxb7-GFP-Tg mouse model to permit visualisation of UB branching morphogenesis in neonatal kidneys, which continue to form nephrons after birth, we previously reported that maternal diabetes may impair UB branching morphogenesis. This ultimately impairs kidney formation [6, 11], resulting in renal dysplasia or hypoplasia, likely via increased cell apoptosis or decreased cell proliferation in the differentiated MM and UB epithelium [6, 11]. The underlying mechanisms appear to be associated with impaired mesenchymal-to-epithelial conversion linked to PAX2 and N-myc interaction [6, 7, 19, 22] and/or excessive nascent nephron apoptosis via activation of NFκB (p50/p65) and p53 pathways [6, 11].
Consistent with our previous studies [6, 11] with the same phenotype in neonatal kidneys, we validated the data obtained from our screening microarray analysis and observed that Hhip expression was significantly increased in the neonatal kidneys of Dia-offspring. Those data led us to speculate whether the augmented HHIP expression induced by high-glucose milieu either directly or indirectly controls SHH-targeting genes involved in cell proliferation, cell fate and cell communication, influencing processes such as epithelial–mesenchymal transition [23, 24]. We further speculated as to whether or not the increased Hhip expression could activate NFκB (p50/p65) and p53 signals and then impact on kidney formation.
During normal kidney development, the balance of HHIP–SHH signalling appears to be critical in the control of a series of sequential events of nephrogenic processes [25, 26]. Upon high glucose insult, for example, in our murine model, the offspring of diabetic dams had a markedly different expression pattern of HHIP when compared with offspring of normal non-diabetic dams. As compared with the limited HHIP expression in Con-offspring (i.e. neonate—UB and MM lineages; age 1 week—collecting duct system), a significant amount of HHIP was detected in the nephrogenic zone of the still developing kidneys of Dia-offspring at the neonatal stage (i.e. secreted HHIP adjacent to UB tips) and at 1 week old (i.e. a nodular expression pattern in glomeruli). These data suggested that HHIP might delay or disorient the usual gradient SHH expression pattern establishment from distal (high) to proximal (low) [25, 26], since HHIP is expressed in cells adjacent to those expressing SHH to antagonise its activity [12]. Our in vitro studies in MK4 and UB tip cells with or without high-glucose media elucidated a potential counterbalance between Hhip and Shh gene expression in both differentiated UB and MM epithelium, and hint that this interaction is critical in the nephrogenic process. Thus, the examination of the functional role of HHIP during the entire period of nephrogenesis with respect to SHH signalling in different renal cell lineages, as well as in the in utero high-glucose environment, merits further investigation.
Alternatively, the heightened HHIP expression might alter the SHH-controlled expression of kidney formation genes, such as Pax2, Sall1, Mycn, Ccnd1, Gli1 and Gli2 [14], in the nephrogenic zone, subsequently resulting in the development of renal abnormalities. For instance, Pax2 as a ‘kidney-specific’ master gene is expressed in both UB and MM lineages, normally optimising UB branching and mesenchymal-to-epithelial transition during kidney development [27–29]. Mutations or deficiency of Pax2 cause an increase in cellular apoptosis [30–32], associated with renal hypoplasia [30, 33, 34]. In the current study, we observed that overexpression of Hhip significantly downregulated genes such as Shh and Pax2; in contrast, overexpression of Pax2 increased Shh and decreased Hhip gene expression in both UB and MK4 cells, underscoring the functional interaction between HHIP, SHH and PAX2.
Thus, we focused on a differentiated MM epithelium (i.e. MK4 cells), since the augmented HHIP expression remained in the developing proximal portion of the nephron in the Dia-offspring, which is mainly derived from differentiated MM lineages. We designed further experiments to elucidate whether the excess amount of HHIP with or without high glucose could modulate the mesenchymal-to-epithelial conversion and nascent nephron apoptosis by studying the MM cell proliferation genes (i.e. Shh, Pax2, Mycn and p27kip1 genes) and apoptosis genes (i.e. genes encoding NFκB [p50/p65] and p53). Indeed, upon exposure to high d-glucose, but not other glucose analogues, there was specific stimulation of Hhip gene expression in a time- and dose-dependent manner. Insulin appears to inhibit the stimulatory effect of high glucose on Hhip gene expression in vitro. Moreover, our current data suggest that PAX2 may be involved in a negative autocrine feedback loop to prevent the high-glucose stimulatory effect on Hhip gene expression by promoting MM cell proliferation genes (i.e. Shh, Pax2, Mycn and p27kip1 genes) and inhibiting MM cell apoptosis genes (i.e. genes encoding NFκB [p50/p65] and p53).
In the current study, we observed that the distribution of HHIP expression is similar to that of TGF-β1 and that TGF-β1 expression was significantly increased in young Dia-offspring with dysmorphogenesis occurring, suggesting that there is functional interaction between HHIP and TGF-β1. It is well-known that TGF-β1 decreases renal branching morphogenesis via its negative feedback mechanisms [35, 36]. Compelling evidence suggests that TGF-β1 plays a key role in glomerular endothelial cell damage [37, 38], production of extracellular matrix and development of renal interstitial fibrosis in diabetic nephropathy [39–41]. In studies in vitro, we further demonstrated that the increase of HHIP expression either by overexpression of Hhip or by the induction of a high-glucose milieu might target TGF-β1 signalling, which, in turn, may lead to events such as phosphorylation of Smad2/3 and decreases in p27Kip1 expression.
In conclusion, the present studies suggest that impaired nephrogenesis induced by maternal diabetes is associated with augmented Hhip gene expression. HHIP targets TGF-β1 signalling, resulting in kidney formation impairment in young Dia-offspring. PAX2 appears to be involved in a negative autocrine feedback loop on Hhip gene expression, attenuating the stimulatory effect of high glucose on Hhip gene expression.
Abbreviations
- Con-offspring:
-
Offspring of non-diabetic dams
- DBA:
-
Dolichos biflorus agglutinin
- DHH:
-
Desert hedgehog
- Dia-offspring:
-
Offspring of diabetic dams
- GFP:
-
Green fluorescent protein
- HHIP:
-
Hedgehog interacting protein
- IHH:
-
Indian hedgehog
- Ins-offspring:
-
Offspring of diabetic dams treated by insulin implantation
- IUGR:
-
Intrauterine growth restriction
- MM:
-
Metanephric mesenchyme
- NFκB:
-
Nuclear factor-kappa B
- PAX2:
-
Paired box gene 2 protein
- qPCR:
-
Quantitative PCR
- SHH:
-
Sonic hedgehog protein
- STZ:
-
Streptozotocin
- Tg:
-
Transgenic
- UB:
-
Ureteric bud
References
Schedl A (2007) Renal abnormalities and their developmental origin. Nat Rev Genet 8:791–802
Yosypiv IV (2011) Renin-angiotensin system in ureteric bud branching morphogenesis: insights into the mechanisms. Pediatr Nephrol 26:1499–1512
Yosypiv IV (2012) Congenital anomalies of the kidney and urinary tract: a genetic disorder? Int J Nephrol 2012:909083
Simeoni U, Ligi I, Buffat C, Boubred F (2011) Adverse consequences of accelerated neonatal growth: cardiovascular and renal issues. Pediatr Nephrol 26:493–508
Amri K, Freund N, Vilar J, Merlet-Benichou C, Lelievre-Pegorier M (1999) Adverse effects of hyperglycemia on kidney development in rats: in vivo and in vitro studies. Diabetes 48:2240–2245
Tran S, Chen YW, Chenier I et al (2008) Maternal diabetes modulates renal morphogenesis in offspring. J Am Soc Nephrol 19:943–952
Zhang SL, Chen YW, Tran S, Chenier I, Hebert MJ, Ingelfinger JR (2007) Reactive oxygen species in the presence of high glucose alter ureteric bud morphogenesis. J Am Soc Nephrol 18:2105–2115
Chugh SS, Wallner EI, Kanwar YS (2003) Renal development in high-glucose ambience and diabetic embryopathy. Semin Nephrol 23:583–592
Kanwar YS, Nayak B, Lin S et al (2005) Hyperglycemia: its imminent effects on mammalian nephrogenesis. Pediatr Nephrol 20:858–866
Chang SY, Chen YW, Zhao XP et al (2012) Catalase prevents maternal diabetes-induced perinatal programming via the Nrf2-HO-1 defense system. Diabetes 61:2565–2574
Chen YW, Chenier I, Chang SY, Tran S, Ingelfinger JR, Zhang SL (2011) High glucose promotes nascent nephron apoptosis via NF-kappaB and p53 pathways. Am J Physiol Renal Physiol 300:F147–F156
Chuang PT, McMahon AP (1999) Vertebrate hedgehog signalling modulated by induction of a hedgehog-binding protein. Nature 397:617–621
St-Jacques B, Hammerschmidt M, McMahon AP (1999) Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev 13:2072–2086
Cain JE, Rosenblum ND (2011) Control of mammalian kidney development by the Hedgehog signaling pathway. Pediatr Nephrol 26:1365–1371
Olsen CL, Hsu PP, Glienke J, Rubanyi GM, Brooks AR (2004) Hedgehog-interacting protein is highly expressed in endothelial cells but down-regulated during angiogenesis and in several human tumors. BMC Cancer 4:43
Zeng X, Goetz JA, Suber LM, Scott WJ Jr, Schreiner CM, Robbins DJ (2001) A freely diffusible form of sonic hedgehog mediates long-range signalling. Nature 411:716–720
Srinivas S, Goldberg MR, Watanabe T, D’Agati V, al-Awqati Q, Costantini F (1999) Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev Genet 24:241–251
Watanabe T, Costantini F (2004) Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev Biol 271:98–108
Chen YW, Liu F, Tran S et al (2006) Reactive oxygen species and nuclear factor-kappa B pathway mediate high glucose-induced Pax-2 gene expression in mouse embryonic mesenchymal epithelial cells and kidney explants. Kidney Int 70:1607–1615
Valerius MT, Patterson LT, Witte DP, Potter SS (2002) Microarray analysis of novel cell lines representing two stages of metanephric mesenchyme differentiation. Mech Dev 112:219–232
Barasch J, Pressler L, Connor J, Malik A (1996) A ureteric bud cell line induces nephrogenesis in two steps by two distinct signals. Am J Physiol 271:F50–F61
Zhang SL, Chen YW, Tran S et al (2007) Pax-2 and N-myc regulate epithelial cell proliferation and apoptosis in a positive autocrine feedback loop. Pediatr Nephrol 22:813–824
Ding H, Zhou D, Hao S et al (2012) Sonic hedgehog signaling mediates epithelial-mesenchymal communication and promotes renal fibrosis. J Am Soc Nephrol 23:801–813
Katoh Y, Katoh M (2009) Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med 9:873–886
Ngan ES, Kim KH, Hui CC (2013) Sonic hedgehog signaling and VACTERL Association. Mol Syndromol 4:32–45
Yu J, Carroll TJ, McMahon AP (2002) Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129:5301–5312
Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M (2002) Nephric lineage specification by Pax2 and Pax8. Genes Dev 16:2958–2970
Dressler GR, Douglass EC (1992) Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc Natl Acad Sci U S A 89:1179–1183
Dressler GR (1996) Pax-2, kidney development, and oncogenesis. Med Pediatr Oncol 27:440–444
Dziarmaga A, Clark P, Stayner C et al (2003) Ureteric bud apoptosis and renal hypoplasia in transgenic PAX2-Bax fetal mice mimics the renal-coloboma syndrome. J Am Soc Nephrol 14:2767–2774
Eccles MR, He S, Legge M et al (2002) PAX genes in development and disease: the role of PAX2 in urogenital tract development. Int J Dev Biol 46:535–544
Porteous S, Torban E, Cho NP et al (2000) Primary renal hypoplasia in humans and mice with PAX2 mutations: evidence of increased apoptosis in fetal kidneys of Pax2(1Neu) +/− mutant mice. Hum Mol Genet 9:1–11
Favor J, Sandulache R, Neuhauser-Klaus A et al (1996) The mouse Pax2(1Neu) mutation is identical to a human PAX2 mutation in a family with renal-coloboma syndrome and results in developmental defects of the brain, ear, eye, and kidney. Proc Natl Acad Sci U S A 93:13870–13875
Maeshima A, Maeshima K, Nojima Y, Kojima I (2002) Involvement of Pax-2 in the action of activin A on tubular cell regeneration. J Am Soc Nephrol 13:2850–2859
Bush KT, Sakurai H, Steer DL et al (2004) TGF-beta superfamily members modulate growth, branching, shaping, and patterning of the ureteric bud. Dev Biol 266:285–298
Rogers SA, Ryan G, Purchio AF, Hammerman MR (1993) Metanephric transforming growth factor-beta 1 regulates nephrogenesis in vitro. Am J Physiol 264:F996–F1002
Brownlee M (2001) Biochemistry and molecular cell biology of diabetic complications. Nature 414:813–820
Brownlee M (2005) The pathobiology of diabetic complications: a unifying mechanism. Diabetes 54:1615–1625
Hills CE, Squires PE (2011) The role of TGF-β and epithelial-to mesenchymal transition in diabetic nephropathy. Cytokine Growth Factor Rev 22:131–139
Hills CE, Siamantouras E, Smith SW, Cockwell P, Liu KK, Squires PE (2012) TGFβ modulates cell-to-cell communication in early epithelial-to-mesenchymal transition. Diabetologia 55:812–824
Lan HY, Chung AC (2012) TGF-β/Smad signaling in kidney disease. Semin Nephrol 32:236–243
Acknowledgements
The authors owe special thanks to J. S. D. Chan (CRCHUM, Montreal, QC, Canada) for his unconditional support and valuable comments on this manuscript.
Funding
This project was supported by grants to SLZ from the Canadian Institutes of Health Research (CIHR, MOP115025) and American Society of Nephrology (ASN)-Carl W. Gottschalk Research Scholar Grant as well as the Bourse de chercheure-boursier Juniors 2-Fonds de recherche en santé du Québec (FRSQ) (SLZ). SYC is a Canadian Diabetes Association Doctoral Student. Editorial assistance was provided by the CRCHUM’s Research Support Office.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
SLZ is the guarantor of this work, had full access to all study data and takes responsibility for data integrity and the accuracy of data analysis. XPZ and SLZ were principal investigators and were responsible for the study conception and design. SLZ drafted, reviewed and edited the manuscript. XPZ, MCL, SYC, SA, YA, IC and SLZ contributed to the in vivo and ex vivo experiments and collection of data. JRI contributed to data discussion and reviewed/edited the manuscript. All authors were involved in the analysis and interpretation of data and contributed to the critical revision of the manuscript. All authors provided final approval for the version to be published.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, XP., Liao, MC., Chang, SY. et al. Maternal diabetes modulates kidney formation in murine progeny: the role of hedgehog interacting protein (HHIP). Diabetologia 57, 1986–1996 (2014). https://doi.org/10.1007/s00125-014-3297-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-014-3297-6