Abstract
Aims/hypothesis
Individuals with low birthweight are at increased risk of type 2 diabetes mellitus. However, the underlying molecular mechanisms are unknown. Previously we have shown that low birthweight is associated with changes in muscle insulin signalling proteins. Here we determined whether low birthweight is associated with changes in insulin signalling proteins in adipose tissue.
Methods
Men (age 23 years) with either a low (bottom 10th percentile) (n = 17) or a normal (50th–90th percentile) (n = 17) birthweight were recruited from the Danish Medical Birth Registry and subcutaneous adipose biopsies were taken.
Results
Between the two groups there was no difference in protein level of the insulin receptor, protein kinase C zeta, glycogen synthase kinase-3 (GSK3) alpha, GSK3 beta, protein kinase B alpha and beta, peroxisome proliferative activated receptor gamma coactivator 1 or Src-homology-2-containing protein. However, the levels of GLUT4 (also known as solute carrier family 2 [facilitated glucose transporter], member 4 [SLC2A4]) (52 ± 10.9% reduction, p < 0.01), p85α subunit of phosphoinositide 3-kinase (PI3K) (45 ± 9% reduction, p < 0.01), p110ß subunit of PI3K (48 ± 17% reduction, p = 0.06) and IRS1 (59 ± 24% reduction, p < 0.05) were reduced in men of low birthweight.
Conclusions/interpretation
These findings show that low birthweight is associated with reduced levels of adipose insulin signalling proteins, thus providing a potential molecular framework to explain why people with low birthweight are at increased risk of developing type 2 diabetes. These differences precede the development of diabetes and thus may help predict disease risk.
Similar content being viewed by others
Introduction
It is well documented that individuals with a low birthweight (LBW) are at increased risk of developing type 2 diabetes mellitus, insulin resistance and other features of the metabolic syndrome in adulthood [1]. The mechanistic basis of this relationship is not understood. However, there is mounting evidence that the early environment may play a critical role. Studies in monozygotic twins have demonstrated that in cases of discordance for type 2 diabetes, the diabetic twin has a significantly lower birthweight than the non-diabetic co-twin [2–4]. Direct evidence for the role of maternal nutrition has been revealed in studies of individuals who were in utero during the Dutch Hunger Winter. These individuals have been shown to have a worse glucose tolerance at age 50 years than those born before the famine [5].
The underlying molecular mechanisms that link poor fetal growth to increased risk of type 2 diabetes many years later are not known. However, we have recently identified a panel of insulin signalling proteins that were present at significantly lower levels in muscle biopsies in young adult LBW men [6]. These included protein kinase Cζ (PKCζ, also known as PRKCZ), GLUT4 (also known as solute carrier family 2 [facilitated glucose transporter], member 4 [SLC2A4]), the p85α subunit of phosphoinositide 3-kinase (PI3K) (p85α, also known as PIK3R1) and the p110ß subunit of PI3K (p110ß, also known as PIK3CB). These protein differences preceded any differences in whole-body insulin sensitivity, suggesting that the former were not secondary consequences of insulin resistance, but could contribute to the increased risk of disease.
Although it has been widely reported that skeletal muscle is the major site of glucose disposal postprandially, recent findings have directed attention towards the major contribution played by adipose tissue. The effect of NEFA on glucose uptake into skeletal muscle has been known for many years [7]. However, the more recent identification of a series of adipokines, including adiponectin and retinol-binding protein 4, has increased the focus on the importance of adipose tissue in whole-body glucose disposal [8, 9]. The importance of adipose tissue in modulating whole-body insulin sensitivity has been further supported by data from transgenic animals. Mice over-producing GLUT4 selectively in adipose tissue show substantially increased glucose tolerance [10]. In addition, animals in which GLUT4 has specifically been knocked out from adipose tissue show insulin resistance in skeletal muscle and liver and glucose intolerance [11].
The aim of the current study was therefore to determine whether young adult LBW men exhibited differential protein levels and mRNA expression in adipose tissue prior to development of whole-body insulin resistance and diabetes.
Subjects and methods
Study protocol and subject characteristics
The study protocol was reviewed and approved by the regional ethical committee and procedures were performed according to the principles of the Helsinki declaration. Written informed consent was obtained from all participants.
As part of our ongoing studies of LBW individuals, we recruited 34 human male volunteers, born between 1979 and 1981 in Copenhagen County, from The Danish National Birth Registry. Recruitment was according to birthweight. Of these men, 17 represented a random subset of a cohort previously described [12], which in addition to undergoing a dual-energy X-ray absorptiometry (DEXA) scan, also underwent a 3-h hyperinsulinaemic–euglycaemic clamp (80 mU m−2 min−1) in combination with indirect calorimetry and a baseline subcutaneous fat biopsy (performed between March and August 2003). As biopsy material was sparse, an additional 17 volunteers were recruited by the same criteria to enable a larger panel of proteins to be studied (studied between July 2004 and March 2005). All measurements and procedures were identical. However, abdominal fat mass was not determined in the second group of subjects. In the first group, nine men had birthweights below the 10th percentile for gestational age (LBW, week 39–41) and eight men had birthweights in the upper normal range (NBW) (50th–90th percentile, week 39–41). In the second group, eight men had an LBW and nine men had an upper NBW. Thus 17 men had birthweights below the 10th percentile for gestational age and 17 had birthweights in the upper normal range. We used the upper NBW range for NBW subjects to maximise both the birthweight differences within the normal distribution, and the chances of observing metabolic and molecular differences between the groups. To avoid confounding from the U-shaped curve, subjects within the highest 10% were excluded [13]. None of the subjects had a family history of diabetes (parents or grandparents) or was receiving medication known to affect glucose or lipid metabolism. Subjects with BMI >30 kg/m2 were excluded. Lifestyle factors such as tobacco use, exercise and diet were not different between groups, as assessed by interview at inclusion. Height, weight, BMI, waist-to-hip ratio and regional DEXA measurements were determined as previously described [12, 14]. Glucose, insulin, NEFA and triacylglycerol were determined at baseline and at 10-min intervals throughout the clamp. Plasma adiponectin was determined at baseline. Estimates of whole-body glucose uptake (M value), glucose and lipid oxidation, and non-oxidative glucose metabolism were obtained at baseline (t = −30 to 0) and during the clamp steady state (t = 150–180 min). An insulin clamp at 80 mU m−2 min−1 was selected to maximise potential differences in insulin sensitivity as our previous study using 40 mU m−2 min−1 had failed to reveal differences in whole-body insulin sensitivity [14]. Blood glucose was determined using a blood glucose meter (Lifescan one touch; Johnson & Johnson, Milpitas, CA, USA). Plasma insulin concentrations were determined by a time-resolved fluorometric immunoassay (AutoDelphia; Perkin Elmer Wallac, Oy, Turku, Finland). Serum NEFA and triacylglycerol were quantified by an in vitro enzymatic colorimetric method (Wako, VA, USA). Plasma adiponectin was quantified by Linco assay (EZHADP-61 K; Electra-box Diagnostica, Denmark).
There were no significant differences in body composition or metabolic characteristics between the first and second batch of subjects.
Fat biopsies
Baseline subcutaneous fat biopsies were obtained from the abdomen during local anaesthesia using a modified Bergstrom needle. Biopsies were blotted free of blood, frozen immediately in liquid nitrogen and stored for later analysis at −80°C.
Chemicals and antibodies
Analytical grade chemicals were obtained from Sigma Chemical or BDH Chemicals (both Poole, Dorset, UK), unless specified otherwise. Rabbit anti-peptide antibodies against glycogen synthase kinase 3 (GSK3) alpha (GSK3A), GSK3 beta (GSK3B), p110β, peroxisome proliferative activated receptor gamma coactivator 1 (PGC1, also known as PPARGC1A), PKCζ and insulin receptor β subunit were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rabbit anti-peptide antibodies against p85α and IRS1 were from Upstate Biotechnology (Lake Placid, NY, USA). The monoclonal antibody to protein kinase B (PKB)α (also known as AKT1) and the rabbit antibodies to Src-homology-2-containing protein (SHC1) and PKBβ (also known as AKT2) were from Cell Signaling (Beverly, MA, USA). The rabbit antibody against GLUT4 was from Chemicon (Temecula, CA, USA).
Protein analysis
Adipose biopsies were extracted in ice-cold solubilisation buffer (50 mmol/l HEPES, 150 mmol/l sodium chloride, 1% Triton X100, 1 mmol/l sodium orthovanadate, 30 mmol/l sodium fluoride, 10 mmol/l sodium pyrophosphate, 10 mmol/l EDTA, 0.5 mmol/l 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 0.4 μmol/l aprotinin, 25 μmol/l bestatin, 12.5 μmol/l E-64A, 10 μmol/l leupeptin hemisulfate, 5 μmol/l pepstatin) using a glass homogeniser. To remove lipid and insoluble material, samples were centrifuged at 15,000 g and at 4°C for 5 min. Protein content of the clarified extracts was determined by a modification of the Lowry method. The protein content of adipose tissue did not differ between any of the groups. Samples were diluted to a common concentration of 0.4 mg/ml in Laemmli buffer and 6 μg total protein subjected to SDS-PAGE. The proteins were transferred to polyvinylidene difluoride membrane (Millipore, MA, USA) and blocked overnight at 4°C (1% milk in Tris-buffered saline with 0.25% Tween 20). Membranes were immunoblotted with antibody against GSK3A, GSK3B, p85α, p110β, PKCζ , insulin receptor β subunit, GLUT4, PKBα, PKBβ, SHC or IRS1 and washed with TBSE (0.1% SDS, 5 mmol/l EDTA, 1% Triton X-100, 1% deoxycholate in Tris-buffered saline), followed by washing in PBS buffer (1% milk in PBS with 0.2% Tween 20). Because of the limited size of the biopsy, samples from the two collection groups were analysed for different proteins to maximise the number of proteins that could be assessed. Thus samples from the first batch of subjects were analysed for GSK3A, GSK3B, p85α, p110β, PKCζ, insulin receptor and GLUT4 and those from the second batch for PKBα, PKBβ, PGC1, SHC (66- and 52-kDa forms) and IRS1. Immunoreactive proteins were detected using horseradish-peroxidase-coupled secondary antibodies and enhanced chemiluminescence reagents according to the manufacturer’s instructions (Amersham, Bucks, UK). ImageQuant software (GE Healthcare, Bucks, UK) was used to quantify specific bands.
For each antibody, control blots were performed, in which varying amounts of protein (5, 10 and 15 μg) were loaded on to the gel to ensure that the enhanced chemiluminescence signal changed in a linear manner. Primary and secondary antibody concentrations were also optimised. In order to ensure the linearity of the signal 10 μg and 5 μg of one sample was loaded on to each gel.
mRNA expression
Total RNA was extracted from frozen subcutaneous adipose tissue using a kit according to the manufacturer’s instructions (Tri Reagent; Sigma-Aldrich, MO, USA). cDNA was generated using a cDNA synthesis kit and random hexamer primers (RevertAid H Minus First Strand; Fermentas Life Sciences, Helsingborg, Sweden). GLUT4, p85α, p110β and IRS1 mRNA were quantified using TaqMan real-time PCR with an ABI 7900 system (Applied Biosystems, CA, USA). Gene-specific probes and primer pairs were used for GLUT4 (Assays-on-Demand, Hs00168966_m1; Applied Biosystems), p85α (Assays-on-Demand, Hs00236128_m1) and p110β (Assays-on-Demand, Hs00927728_m1) and IRS1 (Assays-on-Demand, Hs00178563_m1). For each probe/primer set, a tenfold serial dilution standard curve was generated to confirm linearity with increasing amounts of input cDNA. Each sample was run in duplicate and the transcript quantity calculated according to the standard curve method. The transcript quantity was normalised to the mRNA level of cyclophilin A.
Statistics
All statistical analysis was carried out using Statistica statistical software (StatSoft, Tulsa, OK, USA). The data were normally distributed and there was no significant difference in the variances of the two groups. The significance of any difference between groups was examined by the Student’s t test and a p value of less than 0.05 was considered statistically significant. Results are given as mean ± SEM unless stated otherwise. Correlations between protein levels and mRNA expression and various metabolic and demographic variables were assessed by Spearman’s rank order test.
Before initiation of this study, we considered the issue of statistical power, and calculations were based on our findings of differentially expressed signalling proteins in muscle [6], where n = 20 per group were included and group differences in GLUT4, p85α and p110β were ∼20%, ∼20% and ∼45%, respectively. Assuming differences of similar magnitude in fat, similar standard deviations and a power of 0.80 using a two-tailed test would require 13 individuals per group for GLUT4 , 23 for p85α and seven for p110β. However, our consideration of numbers was also driven by tissue availability, protein yield being much lower from fat biopsies than muscle biopsies. Furthermore, our initial gels run with n = 9 and n = 8 revealed that differences in protein levels were much greater than those observed in muscle. It was thus not justifiable to run further gels using precious and limited amounts of human samples.
Results
Whole-body characteristics
There were no significant differences in any of the parameters measured between the LBW and NBW subjects (Table 1). However, we have previously shown that despite normal whole-body insulin sensitivity, a birthweight in the bottom 10th percentile is associated with slightly increased fasting plasma glucose [12, 14, 15], higher fasting and stimulated plasma insulin concentrations [15], as well as increased abdominal fat mass [12]. Correction of the M value for plasma insulin concentration (i.e. sensitivity index) did not change the data (data not shown). We attribute the lack of differences in the present data set to biological variability in a relatively small cohort.
Protein levels
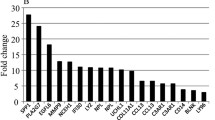
In adipose tissue from the LBW men, protein levels of GLUT4 (p < 0.01), p85α regulatory subunit of PI 3-kinase (p < 0.01), p110ß subunit of PI 3-kinase (p = 0.06) and IRS1 (p < 0.05) were reduced (Fig. 1). However, protein levels of the insulin receptor, PKCζ, GSK3A, GSK3B, PKBα, PKBβ, PGC1 and SHC were similar between the two groups (Table 2).
Insulin signalling protein levels in low birthweight human fat biopsies. Protein levels were determined in human samples by western blotting (see Methods section). Light grey, NBW groups; dark grey, LBW groups. Results are expressed as mean ± SEM. *p < 0.05; **p < 0.01; #p = 0.06
mRNA expression
In contrast to the differences observed at the protein level, there were no differences in mRNA expression of GLUT4, PI3K p85α, PI3K p110β or IRS1 between the NBW and LBW group (Table 3).
Correlations
There was a strong positive correlation between adipose GLUT4 and p85α protein levels (ALL: R = 0.81, p < 0.0001). However, this difference was driven by a correlation in the LBW subjects (LBW: R = 0.70, p = 0.03; NBW: R = −0.11, p = 0.78). No significant correlation between adipose GLUT4 and p110β or between p110β and p85α was found. We detected a negative correlation between GLUT4 protein levels and fasting plasma insulin levels, again primarily driven by a strong correlation in the LBW group (ALL: R = −0.64, p = 0.006; LBW: R = −0.89, p = 0.0001; NBW: R = −0.46, p = 0.26) (Fig. 2a). In addition, GLUT4 correlated positively with whole-body glucose disposal (ALL: R = 0.57, p = 0.03; LBW: R = 0.62, p = 0.086; NBW: R = 0.77, p = 0.10) (Fig. 2b). We have previously reported an association between increased abdominal fat mass and altered fat distribution in young LBW men [12]. Interestingly, in the present study, GLUT4 protein level was negatively associated with abdominal fat mass (ALL: R = −0.46, p = 0.09), but this correlation was seen in the LBW group only (LBW: R = −0.74, p = 0.04; NBW: R = −0.17, p = 0.80).
Discussion
The relationship between LBW and increased risk of diabetes, insulin resistance and other features of the metabolic syndrome was first recognised over a decade ago [1]. However, despite the extensive number of studies that have confirmed this finding, the molecular basis of this relationship remains elusive. There is growing evidence from both human and animal studies that early nutrition may play an important role.
We identified proteins that were at different levels in adipose tissue of young LBW individuals, who, on the basis of their birthweight, have a substantially increased risk of developing type 2 diabetes. but at the time of study displayed no evidence of whole-body insulin resistance or impaired glucose tolerance. In addition they had no family history of diabetes, suggesting that they did not have a genetic susceptibility to the disease. We showed that the levels of a specific panel of insulin signalling proteins were reduced in adipose tissue biopsies from the LBW individuals, namely GLUT4, p85α, p110β, and IRS1. This was a specific effect on this subset of insulin signalling proteins, as levels for a range of other proteins, including the insulin receptor, PKB, GSK, SHC, PKCζ and PGC1, did not differ between the two groups. None of the proteins studied demonstrated parallel differences at the mRNA level. This suggests that the differential protein levels result from post-transcriptional differences. Regulatory mechanisms such as mRNA processing, small interfering RNAs and micro RNAs, together with protein–effector complexes, can control degradation and translation of target transcripts [16]. Interestingly, specific micro RNAs have been shown to regulate insulin secretion [17] and adipocyte differentiation [18].
The dramatic reduction in GLUT4 in adipose tissue from the LBW subjects contrasts with the modest reduction in GLUT4 protein that we observed previously in muscle biopsy samples from similar LBW human subjects [7]. This is consistent with observations by others, demonstrating that, in cases of obesity, insulin resistance and type 2 diabetes, levels of the GLUT4 glucose transporter are decreased selectively in adipocytes [19]. In the present study, adipose tissue GLUT4 content was negatively correlated with fasting plasma insulin concentrations and was positively correlated with whole-body glucose disposal. This provides further evidence that adipose tissue GLUT4 plays a key role in regulation of glucose disposal by tissues such as muscle. The precise mechanism by which this occurs is not known. However, a number adipokines have been identified [9]. Plasma adiponectin levels did not differ significantly between the two groups, suggesting that it may not be the link in this instance.
The protein level of IRS1 was also greatly reduced in adipose tissue from LBW individuals. It is known that IRS1 is reduced in adipose tissue from type 2 diabetic patients [20], and in healthy first-degree relatives of subjects with type 2 diabetes [21]. This suggests that the reduction in IRS1 is not a consequence of persistent hyperglycaemia or hyperinsulinaemia, but represents a very early defect that predisposes a person to type 2 diabetes.
The reduction in p110ß protein level was consistent with our previous findings in muscle biopsy samples, which indicated that adipose tissue within those biopsies may also be a target of fetal programming [7]. In addition, we have also shown previously that adipose tissue p110ß levels are dramatically reduced in a rodent model of nutritionally-induced in utero growth restriction and type 2 diabetes [22]. The reduction in p85α catalytic subunit of PI3K was unexpected. It has been reported that there is no difference in p85α in adipose tissue from type 2 diabetic patients compared with control individuals [23]. However, this lack of difference may be due to a compensatory increase of expression in diabetic patients, in response to the diabetic metabolic environment.
The LBW subjects in this study did not demonstrate a statistically significant reduction in insulin action as determined by the gold standard hyperinsulinaemic–euglycaemic clamp technique. We have previously found evidence of modest reductions in insulin stimulated whole-body glycolysis [14] and muscle (forearm) glucose uptake [24] in a similar population of Danish LBW men. The reduction in insulin signalling components was much more pronounced than the degree of insulin resistance in the LBW individuals. The observation that these differences are present in these otherwise healthy individuals with normal glucose tolerance suggests that such differences are important primary programmed abnormalities, which could link an adverse intrauterine environment to increased risk of diabetes. These defects must be compensated for by other unknown mechanisms at this early stage in the pathogenesis of disease. They could still play a key role in facilitating the development of overt insulin resistance and type 2 diabetes, when the added impact of age, physical inactivity and/or obesity are present.
For obvious reasons we have no way of knowing which of these subjects will develop diabetes. However, we were encouraged by the strong correlation between GLUT4 and measures of body fat (e.g. abdominal fat) in the LBW subjects. LBW has been shown to be associated with later obesity [25], and the combination of LBW and later obesity appears particularly detrimental in terms of diabetes risk [26].
In summary, these findings demonstrate that LBW individuals have substantially reduced levels of key insulin signalling proteins in adipocytes, thus providing a potential molecular framework to explain why such individuals are at increased risk of developing type 2 diabetes. These differences are present before overt diabetes develops and may therefore represent early markers of disease risk. There is growing evidence that the early environment plays a key role in mediating the relationship between poor early growth and type 2 diabetes. In light of the rapidly increasing number of cases of diabetes worldwide, it is critical to understand such an environmentally driven process, since it provides a much more suitable target for intervention than those resulting from genetic alterations.
Abbreviations
- GSK3:
-
glycogen synthase kinase 3
- GSK3A:
-
glycogen synthase kinase 3 alpha
- GSK3B:
-
glycogen synthase kinase 3 beta
- LBW:
-
low birthweight
- NBW:
-
normal birthweight
- p85α:
-
the p85α subunit of phosphoinositide 3-kinase (also known as PIK3R1)
- p110ß:
-
the p110β subunit of phosphoinositide 3-kinase (also known as PIK3CB)
- PGC1:
-
peroxisome proliferative activated receptor gamma coactivator 1 (also known as PPARGC1A)
- PI3K:
-
phosphoinositide 3-kinase
- PKB:
-
protein kinase beta
- PKCζ :
-
protein kinase Cζ (now known as PRKCZ)
- SHC:
-
Src-homology-2-containing protein
References
Hales CN, Barker DJ (2001) The thrifty phenotype hypothesis. Br Med Bull 60:5–20
Poulsen P, Vaag AA, Kyvik KO, Moller Jensen D, Beck-Nielsen H (1997) Low birthweight is associated with NIDDM in discordant monozygotic and dizygotic twin pairs. Diabetologia 40:439–446
Bo S, Cavallo-Perin P, Scaglione L, Ciccone G, Pagano G (2000) Low birthweight and genetic abnormalities in twins with increased susceptibility to type 2 diabetes mellitus. Diabet Med 17:365–370
Iliadou A, Cnattingius S, Lichtenstein P (2004) Low birthweight and type 2 diabetes: a study on 11162 Swedish twins. Int J Epidemiol 33:953–954
Ravelli AC, Van der Meulen JH, Michels RP et al (1998) Glucose intolerance in adults after prenatal exposure to famine. Lancet 351:173–177
Ozanne SE, Jensen CB, Tingey KJ, Storgaard H, Madsbad S, Vaag AA (2005) Low birthweight is associated with specific changes in muscle insulin-signalling protein expression. Diabetologia 48:547–552
Randle PJ, Garland PB, Hales CN, Newsholme EA (1963) The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet 1:785–789
Graham TE, Yang Q, Bluher M et al (2006) Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Eng J Med 354:2552–2563
Rajala MW, Scherer PE (2003) Mini review: the adipocyte at the crossroads of energy homeostasis, inflammation and atherosclerosis. Endocrinology 144:3765–3773
Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB (1993) Adipose tissue hyperplasia and enhanced glucose disposal in transgenic mice over-expressing GLUT 4 selectively in adipose tissue. J Biol Chem 268:22243–22246
Abel ED, Peroni O, Kim JK et al (2001) Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 409:729–733
Rasmussen EL, Malis C, Jensen CB et al (2005) Altered fat distribution in young adult men who had low birthweight. Diabetes Care 28(1):151–153
McCance DR, Pettitt DJ, Hanson RL, Jacobsson LT, Knowler WC, Bennett PH (1994) Birthweight and non-insulin dependent diabetes: thrifty genotype, thrifty phenotype or surviving small baby genotype? Br Med J 308:942–945
Jensen CB, Storgaard H, Dela F, Holst JJ, Madsbad S, Vaag AA (2002) Early differential defects of insulin secretion and action in 19-year old Caucasian men who had low birthweight. Diabetes 51:1271–1280
Schou JH, Pilgaard K, Vilsboll T et al (2005) Normal secretion and action of the gut incretin-like peptide-1 and glucose dependent insulinotropic polypeptide in young men with low birthweight. J Clin Endocrinol Metab 90:4912–4919
Tomari Y, Zamore PD (2005) Perspective: machines for RNAi. Genes Dev 19:517–519
Poy MN, Eliasson L, Krutzfeldt J et al (2004) A pancreatic islet-specific microRNA regulates insulin secretion. Nature 423:226–230
Esau C, Kang X, Peralta E et al (2004) Micro RNA-143 regulates adipocyte differentiation. J Biol Chem 279:52361–52365
Shepherd PR, Kahn BB (1999) Glucose transporters and insulin action — implications for insulin resistance and diabetes mellitus. N Eng J Med 341:248–257
Rondinone CM, Wang LM, Lonnroth P, Wesslau C, Pierce JH, Smith U (1997) Insulin receptor substrate (IRS) 1 is reduced and IRS-2 is the main docking protein for phosphatidylinositol 3-kinase in adipocytes from subjects with non-insulin-dependent diabetes mellitus. Proc Natl Acad Sci 94:4171–4175
Smith U, Axelson M, Carvalho E, Eliasson B, Jansson PA, Wesslau C (1999) Insulin signalling and action in fat cells: associations with insulin resistance and type 2 diabetes. Ann N Y Acad Sci 892:119–126
Ozanne SE, Nave BT, Wang CL, Shepherd PR, Prins J, Smith GD (1997) Poor fetal nutrition causes long-term changes in expression of insulin signalling components in adipocytes. Am J Physiol Endocrinol Metab 273:E46–E51
Rondinone CM, Carvalho E, Wesslau C, Smith UP (1999) Impaired glucose transport and protein kinase B activation by insulin, but not okadaic acid in adipocytes from subjects with type 2 diabetes mellitus. Diabetologia 42:819–825
Hermann TS, Rask-Madsen C, Ihlemann N et al (2003) Normal insulin-stimulated endothelial function and impaired insulin-stimulated muscle glucose uptake in young adults with low birthweight. J Clin Endocrinol Metab 88:1252–1257
Law CM, Barker DJ, Osmond C, Fall CH, Simmonds SJ (1992) Early growth and abdominal fatness in adult life. J Epidemiol Community Health 46:184–186
Hales CN, Barker DJ, Clark PM et al (1991) Fetal and infant growth and impaired glucose tolerance at age 64. Br Med J 303:1019–1022
Acknowledgements
We thank M. Modest and S. Riemer for expert technical assistance. We are grateful to N. Hales for many helpful discussions. This work was supported by the British Heart Foundation, the Parthenon Trust, the National Institutes of Health, the Danish Diabetes Association and the Novo Nordisk Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ozanne, S.E., Jensen, C.B., Tingey, K.J. et al. Decreased protein levels of key insulin signalling molecules in adipose tissue from young men with a low birthweight – potential link to increased risk of diabetes?. Diabetologia 49, 2993–2999 (2006). https://doi.org/10.1007/s00125-006-0466-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-006-0466-2