Abstract
Key message
We identified and characterized a mutant of soybean stachyose synthase gene controlling reduced stachyose content which benefit the soybean seed composition breeding program in the future.
Abstract
It has been shown that in soybean, increased sucrose and reduced raffinose family oligosaccharides would have a positive impact on the world’s feed industry by improving digestibility and feed efficiency. We searched for new sources of modified oligosaccharide content in a subset of the USDA Soybean Germplasm Collection and then identified plant introduction (PI) 603176A as having ultra-low stachyose content (0.5 %). We identified a 33-bp deletion mutant in the putative stachyose synthase gene (STS gene, Glyma19g40550) of PI 603176A. A co-dominate indel marker was successfully developed from this 33-bp deletion area and was genetically mapped into two F 2:3 populations and a F 4:5 population, which associated with low stachyose content in the progeny lines. These observations provided strong evidence that the STS gene is responsible for stachyose biosynthesis in the soybean plant. Expression of the sts gene remained at the normal level, suggesting the loss of function in the gene is due to defective protein function. This gene-based perfect genetic marker for low stachyose content can be useful for marker-assisted selection in soybean molecular breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean seed is a major food source providing protein, oil, carbohydrates, secondary metabolites, and other nutrients to humans and animals. The seed is comprised of on average 40 % protein, 20 % oil, and 33 % carbohydrates, of which up to 16.6 % of the total carbohydrates are soluble sugars (Hymowitz and Collins 1974). The major components of the soluble sugars are glucose, fructose, sucrose, raffinose, and stachyose. The amount of major soluble sugar components among soybean germplasm varies; e.g. sucrose 1.5–10.2 %, stachyose 1.4–6.7 %, and raffinose 0.1–2.1 % of the total dry matter (Hou et al. 2009; Hymowitz and Collins 1974).
Raffinose is a trisaccharide that can be found in the cotyledons, seed coats, and hypocotyls (Bentsink et al. 2000). Stachyose is a tetrasaccharide, which is recognized as an important transport carbohydrate in a large number of woody plants, cucurbits and legumes (Peterbauer et al. 1999). The most common raffinose family oligosaccharides (RFOs) are trisaccharide raffinose, tetrasaccharide stachyose and the pentasaccharide verbascose (Minorsky 2003). RFOs can act as reserve carbohydrates, membrane stabilizers and stress tolerance mediators (Bentsink et al. 2000; Elsayed et al. 2013; Karner et al. 2004; Van den Ende 2013). Since soybean meal is a common source of protein for livestock (Meis et al. 2003) and humans (Guimaraes et al. 2001), soybean digestibility is important. Higher concentration of RFOs in seeds is one of the major problems in the efficient utilization of soybean for human food and animal feed applications. In soy meal fed monogastric animals poor digestibility of RFOs causes a reduction in metabolizable energy and an increase in flatulence and diarrhea. In addition to their indigestibility, raffinose and stachyose can cause diarrhea that may increase digesta passage rate and decreased digestion and absorption of dietary nutrients (Parsons et al. 2000).
Specific galactosyltransferases enzymes (e.g. raffinose synthase, stachyose synthase, etc.) catalyze the reaction towards biosynthesis of raffinose and stachyose from sucrose. A number of gene sequences have been annotated as raffinose synthases, but biochemical confirmation and molecular characterization have only been completed for maize (Zhou et al. 2012), pea (Peterbauer et al. 1999), soybean (Dierking and Bilyeu 2008; Skoneczka et al. 2009) and rice (Li et al. 2007). In Arabidopsis thaliana, a knock-out mutation of raffinose synthase (RS) or the overexpression of galactinol synthase (GolS) caused the reduction of leaf raffinose levels when compared to wild type (Zuther et al. 2004). To our knowledge, there was no evidence in the literature that stachyose synthase exists in soybean. The only published experiments indicate that an adzuki bean stachyose synthase is capable of catalyzing the synthesis of both stachyose and verbascose (Peterbauer et al. 1999, 2002). Overexpression of STS from adzuki bean (Vigna angularis) in Arabidopsis had accumulated stachyose upon cold acclimation (Iftime et al. 2011).
The reduction in oligosaccharide levels in soybean meal could increase the amount of soy proteins in rations (Hartwig et al. 1997). In past years, many efforts have been made to evaluate existing soybean germplasm and mutagenized materials aiming for the improvement of digestible carbohydrates and better nutritional factors. Sebastian et al. (2000) screened bulk seed from approximately 8000 individual M3 generation plants and the USDA Soybean Germplasm Collection, and identified two types of modified carbohydrate profile soybean seeds. Soybean accession PI 200508 was identified as having reduced levels of RFOs and elevated levels of sucrose (Kerr and Sebastian 2000). Initial characterization of this PI was carried out by Hitz et al. (2002). Later, it was reported that PI 200508 allele of RS2 (raffinose synthase) was associated with the increased sucrose and low raffinose and stachyose seed phenotype (Dierking and Bilyeu 2008; Skoneczka et al. 2009). The intellectual property of the above soybean line limits the development of these traits in public breeding programs.
In an effort to discover new sources of modified sucrose and RFO’s, a subset of over 650 soybean germplasm accessions with maturity group (MG) ranging from III to V were evaluated at the University of Missouri. This initial screening has identified several potential PIs with lower raffinose and stachyose content and higher sucrose level. These PIs were evaluated for sugar composition stability in different growing environments. Here we report the identification of a stachyose synthase gene controlling reduced stachyose content in soybean PI 603176A (0.5 %), from which a co-dominant genetic marker was successfully developed and genetically mapped. This gene-based genetic marker can be useful in molecular soybean breeding programs, aiming towards breeding of modification of sucrose and RFOs in soybean.
Materials and methods
Screening of seed raffinose and stachyose content in 650 soybean germplasm
A total of 650 soybean accessions (from MG III to V) were requested from GRIN (Germplasm Resources Information Network) (http://www.ars-grin.gov). The seed raffinose and stachyose content of these 650 accessions were quantified by standard HPLC method which was described at the below “Sugar content quantification of soybean seeds by HPLC”.
Population development
Two newly identified soybean accessions with low RFOs (stachyose + raffinose), PI 603176A and PI 594012, along with elite lines with regular RFO’s content, S07-5049 and S05-11482, were utilized to develop genetic populations in this study. A cross of line S07-5049 and PI 603176A was made in the summer of 2010 at the Bradford Research and Extension Center (BREC), University of Missouri, Columbia, MO. F 1 hybrid of this cross were planted in Costa Rica and F 2 seeds were planted at BREC in summer of 2011. One hundred and thirty-one resulting F 2:3 progenies derived from this cross were screened for seed oligosaccharide content using HPLC system (Hou et al. 2009). A second population of 70 F 2:3 progenies was developed from a PI 594012 × PI 603176A cross. In addition, a third population of 82 F 4:5 advanced inbred lines developed from a S05-11482 × PI 603176A cross was subsequently employed for confirmation tests. Both these two populations were planted in Costa Rica and seeds were shipped back for further analysis.
DNA extraction and sequencing of stachyose synthases gene (STS gene)
Genomic DNA was isolated from young leaf tissue of the parents and progeny plants using a standard CTAB protocol (Vuong et al. 2010). The DNA concentration was quantified with a spectrophotometer (NanoDrop Technologies Inc., Centreville, DE) and diluted to a concentration of 50 ng/µl for polymerase chain reaction (PCR) amplification to amplify the target regions or for sequencing.
Soybean sequences were obtained after PCR and either direct sequencing or cloning followed by sequencing (Wu et al. 2010). PI 603176A, S07-5049, and their progeny lines carrying the STS alleles were sequenced from PCR products, which were amplified with primers that were intronic, flanking exonic sequences.
Allele-specific molecular marker assay development
PCR amplifications were performed in 25 µl final volume on the Eppendorf 96-well thermal cyclers with three primers: Primer_GCF_G:GCGGGCAGGGCGGCAGGGTGATGGGAGATTCCTTG, Primer_GCF_T:GCGGGCAGGGTGATGGGAGATTCCTTT, Primer_common: ACTCAAAAGCAACATCAGAACCAT (Eppendorf AG, Germany). Each reaction contained 40–50 ng of genomic DNA, 0.13 µM of forward primer and reverse primer, 0.2 mM of each dNTP, and SYBRGreen mix solution (GenScript Corp., Piscataway, NJ). The thermal cycler program was performed at 95 °C for 5 min followed by 35 cycles of 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s. Melting curve from 60 to 85 °C, with readings taken every 0.1 °C.
Sugar content quantification of soybean seeds by HPLC
Seed sucrose and RFO contents of soybean lines in each genetic population were quantified. Approximately 1 g of dried seed from each line was ground to fine powder. 0.1 g ground soybean powder was air-dried for 2 days, followed by the addition of 0.9 ml of HPLC grade water in a 2-ml centrifuge vial. Sample tubes were incubated at 55 °C for 20 min with 200 rpm agitation. Subsequently, 0.9 ml of 95 % acetonitrile was added and vortexed for 30 s, followed by centrifuging for 10 min, and filtered with a syringe and 0.45 µm filter. A sample solution of 100 µL was mixed with 400 µL of 65 % acetonitrile in an HPLC vial. Standard sugar melibiose (Sigma Chemical Co., St. Louis, MO) was used as the internal standard. A soybean cultivar, Williams 82, and a low-stachyose line, PI 200508, were included as checks in each extraction and quantification in order to monitor the consistency and accuracy of the tests.
Expression analysis by quantitative RT-PCR
Primer sequences for the candidate genes are STS: 5′-GGGTGATGGGAGATTCC-3′ and 5′-CTCAAAAGCAACATCAGAACC-3.
The primers for the housekeeping gene, elongation factor 1α are 5′-CTGTAACAAGATGGATGCCACTAC-3′ and 5′-CAGTCAAGGTTAGTGGACCT-3′ (Czechowski et al. 2005). The real time polymerase chain reaction (RT-PCR) was performed using the QuantiTect SYBR Green RT-PCR Kit (Qiagen, Valencia, CA) in 10 μL reactions. The parameters for the one step RT and the PCR were as follows: reverse transcription at 50 °C for 30 min followed by 95 °C for 15 min, then 35 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s with an ending hold at 4 °C. Experiments included control reactions lacking the reverse transcriptase enzyme to assess possible genomic DNA contamination.
Results
Screening of seed raffinose and stachyose content in the soybean germplasm
A total of 650 soybean accessions (from MG III to V) were screened for natural variation of seed sugar components. The frequency distribution of seed raffinose and stachyose content for 650 accessions are shown in Supplement S1. The initial screening identified a few lines with low raffinose and low stachyose content. Those lines were planted in different environments for the stability test. Subsequently, the lines with low raffinose and low stachyose content in more than two environments were selected for further genetic analysis (Table 1). PI 603176A showed a significant reduction in stachyose content from 5 to 0.5 % in different environments. Three genetic populations derived from this PI line were developed for genetic analysis and gene mapping.
Identification of the STS gene mutation in PI 603176A and single nucleotide polymorphism (SNP) variation in the soybean germplasm.
The stachyose synthase gene (STS, Glyma19g40550) is a putative gene involved in a pathway which converts raffinose to stachyose. Besides that, the genes RS2 (Glyma06g18890), RS3 (Glyma05g08950), and RS4 (Glyma05g02510) were confirmed to be involved with the raffinose metabolism. We have sequenced these four genes in PI 603176A along with other lines (Table 1). A number of SNP variations were identified in these four genes (Table 1). Interestingly, the sequence of the STS gene in PI 603176A showed that there is a 33-bp deletion in the exon 4 of this gene compared to an elite line S07-5049, which caused the 11 amino acid deletion in the protein (Fig. 1).
We also identified new non-synonymous SNPs, such as G2000T and C2123A in gene RS2, C197G in gene RS4, T1895A and T2558G in STS gene, which caused the amino acid changes (Table 1). The PI 594012 showed second lowest stachyose content at 2.89 % among the 650 germplasm accessions evaluated in this study. There were three non-synonymous SNPs in the RS2 and RS3 genes and one non-synonymous SNP in STS gene of PI 594012. In addition, two other PI lines, PI 424079 and PI 437654, also showed the lowest raffinose content with the various non-synonymous SNPs in these four genes. Further genetic analysis is needed for the functional confirmation of these SNP variation.
Statistical analysis of seed stachyose content in a genetic population
A new cross of PI 603176A and an elite soybean line S07-5049 with regular raffinose and stachyose content was made to produce an F 2:3 population. PI 603176A has high raffinose content (3.31 %) and low stachyose content (0.55 %), while S07-5049 has regular raffinose (0.9 %) and stachyose content (4.6 %) (Table 2; Fig. 2). We have analyzed an F 2:3 population derived from a cross between these two parental lines. The stachyose and raffinose content showed characterization of a Mendelian factor with a segregation ratio of 1:2:1 (P > 0.1). A significant negative correlation (r 2 = −0.92, P < 0.001) was observed between raffinose and stachyose content in this genetic population (Table 3). In contrast, no significant correlation was found between sucrose content and raffinose/stachyose content in this population.
Scatterplot of percent seed sucrose and raffinose content (Y-axis) and percent seed stachyose content (X-axis) in Population 1 (S07-5049 × PI 603176A, a, c) and Population 2 (PI 594012 × PI 603176A, b, d). Sucrose values are represented by triangles and raffinose values by diamonds. Trend line equations are given at the top right of each plot
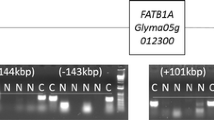
The identification of the STS gene
Statistical analysis of phenotypic data in the S07-5049 × PI 603176A population showed that a single gene plays key role in a pathway of raffinose to stachyose. We developed this deletion (sts genotype) into the Allele-Specific molecular marker assay (Fig. 3). The marker assay data from F 2:3 plants for both candidate genes and data sets revealed 1:2:1 ratio (P < 0.05) for PI 603176A alleles (sts/sts): heterozygote (STS/sts): S07-5049 alleles (STS/STS) (Table 1). The association analysis of phenotypic data of raffinose and stachyose content and genotypic data determined a strong association of the RFOs content and the sts genotype. A significant positive correlation (r 2 = 0.89, P < 0.001) was observed between stachyose content and the sts genotype. In contrast, a significant negative correlation (r 2 = −0.86, P < 0.001) was observed between raffinose content and the sts genotype. Results confirmed the biochemical function of STS gene in converting raffinose into stachyose. The functional STS gene in S07-5049 converts raffinose into stachyose and causes low raffinose and high stachyose content in regular soybean seeds. The sts mutant in PI 603176A resulted in the seed’s inability to convert raffinose to stachyose and the accumulation of raffinose content, which as same as in the progeny lines.
Expression analysis of STS gene by quantitative RT-PCR
Expression of the STS gene was determined using quantitative RT-PCR in cv. Williams 82, S07-5049, and PI 603176A. Leaf tissue from these three parental lines was collected at the vegetative stage V1, reproductive stages R1, and R5 growth stages and seed tissue was collected at the R5 and R6 growth stages (Fehr and Caviness 1977). The results indicated that the putative stachyose synthase genes have similar transcript levels in the three parental lines when compared to the housekeeping gene, elongation factor 1α (Czechowski et al. 2005). There is a slight increase of expression in sts genes of PI 603176A from the V1 to R6 growth stage (Fig. 4). Both STS and sts genes do not appear to be tissue specific or highly expressed in the developing seed tissues. Similar transcript levels as determined by quantitative RT-PCR, for STS and sts, and for all three lines, indicated that the difference in oligosaccharide content was unlikely to be a product of disruption in the transcriptional machinery, but rather a functional loss of protein.
Analyzing oligosaccharide content in the two other populations and progeny lines
In order to test the stability of the sts mutant across the generation and environments, we selected three low stachyose F 2:3 progenies for breeding purpose. Fourteen F 4 lines were harvested in a greenhouse in May, 2013. The sequences and the genetic markers had confirmed the sts genotype in these 14 lines. Sugar analysis revealed that these 14 lines had low stachyose content (~0.5 %).
Moreover, over 70 F 2:3 lines of the PI 594012 × PI 603176A population and 82 F 4:5 lines of the S05-11482 × PI 603176A population were developed for the confirmation test. The phenotypic data and genotypic data were matched perfectly within progeny lines in these two populations (Table 2). The sts genotype is always associated with low stachyose content and high raffinose content.
Phylogenetic relationships of different RS and STS proteins in plant genomes
Both RS and STS genes contain the same Pfam structure, PF05691 (Raffinose_synthase). Based on the EMBI (European Bioinformatics Institute) data base, 136 Eukaryote, 53 Bacteria and 21 Archaea species shared the PF05691 structure domain. About 101 RS and STS protein sequences from 43 plant genomes were selected to generate phylogenetic tree (Fig. 5). The soybean STS genes shared highest amino acid similarity with STS genes compared to species Phaseolus vulgaris and Vigna angularis at 87 %, following a similarity of 78 % in Cicer arietinum and Medicago truncatula. It is not surprising that the soybean RS and STS genes were grouped into different subclades due to only 45 % amino acid similarities between them. The sequence identity for soybean RS2 and RS3 is 65 %, for RS2 and RS4 is 70 %, and for RS3 and RS4 it is 62 % at the amino acid level, which is shown in the phylogenetic tree (Fig. 5).
Discussion
We have successfully identified a 33-bp deletion mutant in the putative soybean stachyose synthase (STS) gene, characterized their expression in multiple tissues, developed and characterized the phenotype–genotype association in three segregating genetic populations. We confirmed the function of STS gene, which can convert raffinose into stachyose as part of raffinose metabolism (Dierking and Bilyeu 2008; Skoneczka et al. 2009). The sts mutant cut off the conversion of raffinose to stachyose, which caused a 90 % reduction of stachyose and the accumulation of raffinose content in soybean seeds. These data provided strong evidence that the STS gene was responsible for stachyose biosynthesis in the soybean plant. The sts genotype was always associated with low stachyose content (0.5 %), showing a significant reduction from regular stachyose content (5 %). This sts genotype mutant from PI 603176A associated with the low seed stachyose content was also confirmed in different progeny lines from three genetic populations which under different growing environments.
Raffinose and stachyose are tri-saccharides and tetra-saccharides, respectively. These two types of enzymes are involved mainly in the biosynthesis of RFOs. The raffinose synthase is the galactinol synthase, which generates galactinol from galactose and myo-inositol, the second one being galactosyltransferases (raffinose and stachyose synthases), which are responsible for incorporating the galactosyl group from galactinol into the oligosaccharide (Peterbauer et al. 1999, 2002). The STS sequences in adzuki bean, maize, and soybean share a high similarity with known RS sequences because both RS and STS contain the same Pfam structure (Raffinose_synthase, PF05691) (Peterbauer et al. 1999; Zhou et al. 2012). It is also notable that based on the EMBI database, 136 Eukaryote, 53 Bacteria, and 21 Archaea species shared the PF05691 structure domain, which might indicate the evolutionary history of the raffinose synthase gene. Nevertheless, the adzuki bean stachyose synthases are distinguishable from raffinose synthases by a characteristic central insertion of 70–80 amino acids (Peterbauer et al. 1999, 2002). Likewise, the amino acid sequence identity is 45 % for STS and RS genes in soybean and 42 % in pea (Dierking and Bilyeu 2008). The blast hit result from NCBI database shows that the STS genes in Phaseolus vulgaris and Vigna angularis have the highest amino acid similarity at 87 % when compared with soybean. This is followed by a 78 % similarity in Cicer arietinum and Medicago truncatula, which suggests that the STS genes in these species might have the same stachyose synthase function.
So far, the RS2 mutant allele from PI 200508 was the main source for the selection of low raffinose and stachyose seed phenotype in soybean (Dierking and Bilyeu 2008). Typically, the seed raffinose content of the RS2 allele remains at a regular level of 1 %, but the stachyose content drops to 1.5 %. It was reported that a soybean line, 222-18-1, exhibited ultra-low raffinose content (0.13 %) and stachyose content (0.58 %), and in which the RS2 allele was combined with other RS3 and RS4 mutant alleles (Schillinger et al. 2011). This might suggest the RS2, RS3, and RS4 alleles are three main enzymes responsible for converting galactinol into raffinose; however, the function of RS3 and RS4 are still unknown. The sts mutant allele in our study causes only at a level of 0.5 % stachyose content in seed. The sts genes are highly expressed in both leaf and developing seeds, which indicated that the 11-amino acids deletion might cause a functional loss of protein. There was still a production of 0.5 % stachyose content in the mutant. We speculate that this was possibly due to a partial function of the sts mutant or from the functional alleles of RS3 and RS4.
Due to the increased use of soybeans and soy products in the feed industry, it is important to understand the nutritional and anti-nutritional components of soybean. Soy meal fed monogastric animals suffer from a reduction in metabolizable energy and an increase in flatulence and diarrhea (Coon et al. 1990). The sts allele was always associated with a low stachyose content (0.5 %), but at the same time it was associated with a high raffinose content (4.5 %) which preserves the total RFOs content at a regular level. Although the best combination level of raffinose/stachyose content for animal feeding has not been well determined, the sts allele can be combined with other RS2/RS3/RS4 alleles to achieve a more nutritional combination of raffinose/stachyose content in soybean seeds. Molecular marker assays associated with the 90 % of stachyose content reduction were developed and validated in three mapping populations throughout different environments. This genetic marker will allow the incorporation of the desirable RFO traits into soybean varieties through molecular breeding.
Author contribution statement
D.Q, B.V and T.D.V designed research; D.Q, B.V, T.D.V, B.G, H.S and J.G.S performed research; D.Q analyzed data and wrote the manuscript; T.D.V, B.V and H.T.N edited the manuscript; and H.T.N oversaw the project.
References
Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SP, Koornneef M (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124:1595–1604
Coon CN, Leske KL, Akavanichan O, Cheng TK (1990) Effect of oligosaccharide-free soybean meal on true metabolizable energy and fiber digestion in adult roosters. Poult Sci 69:787–793
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dierking EC, Bilyeu KD (2008) Association of a soybean raffinose synthase gene with low raffinose and stachyose seed phenotype. Plant Genome J 1:135
Elsayed AI, Rafudeen MS, Golldack D (2013) Physiological aspects of raffinose family oligosaccharides in plants: protection against abiotic stress. Plant Biol 16:1–8
Fehr WR, Caviness CE (1977) Stages of soybean development. Coop Ext Serv Spec Rep 80 Iowa State Univ, Ames
Guimaraes VM, de Rezende ST, Moreira MA, de Barros EG, Felix CR (2001) Characterization of alpha-galactosidases from germinating soybean seed and their use for hydrolysis of oligosaccharides. Phytochemistry 58:67–73
Hartwig EE, Kuo TM, Kenty MM (1997) Seed protein and its relationship to soluble sugars in soybean. Crop Sci 37:770–773
Hitz WD, Carlson TJ, Kerr PS, Sebastian SA (2002) Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype on soybean seeds. Plant Physiol 128:650–660
Hou A, Chen P, Alloatti J, Li D, Mozzoni L, Zhang B, Shi A (2009) Genetic variability of seed sugar content in worldwide soybean germplasm collections. Crop Sci 49:903
Hymowitz T, Collins FI (1974) Variability of sugar content in seed of Glycine max (L.) Merril and G. soja Sieb and Zucc. Agron J 64:613–616
Iftime D, Hannah MA, Peterbauer T, Heyer AG (2011) Stachyose in the cytosol does not influence freezing tolerance of transgenic Arabidopsis expressing stachyose synthase from adzuki bean. Plant Sci Int J Exp Plant Biol 180:24–30
Karner U, Peterbauer T, Raboy V, Jones DA, Hedley CL, Richter A (2004) myo-Inositol and sucrose concentrations affect the accumulation of raffinose family oligosaccharides in seeds. J Exp Bot 55:1981–1987
Kerr PS, Sebastian A (2000) Soybean products with improved carbohydrate composition and soybean plants. US Patent 6147193 A
Li S, Kim WD, Kaneko S, Prema PA, Nakajima M, Kobayashi H (2007) Expression of rice (Oryza sativa L. var. Nipponbare) alpha-galactosidase genes in Escherichia coli and characterization. Biosci Biotechnol Biochem 71:520–526
Meis SJ, Fehr WR, Schnebly SR (2003) Seed source effect on field emergence of soybean lines with reduced phytate and raffinose saccharides. Crop Sci 43:1336–1339
Minorsky PV (2003) The hot and the classic. Plant Physiol 132:25–26
Parsons CM, Zhang Y, Araba M (2000) Nutritional evaluation of soybean meals varying in oligosaccharide content. Poult Sci 79:1127–1131
Peterbauer T, Mucha J, Mayer U, Popp M, Glossl J, Richter A (1999) Stachyose synthesis in seeds of adzuki bean (Vigna angularis): molecular cloning and functional expression of stachyose synthase. Plant J Cell Mol Biol 20:509–518
Peterbauer T, Mach L, Mucha J, Richter A (2002) Functional expression of a cDNA encoding pea (Pisum sativum L.) raffinose synthase, partial purification of the enzyme from maturing seeds, and steady-state kinetic analysis of raffinose synthesis. Planta 215:839–846
Schillinger JA, Dierking EC, Bilyeu KD (2011) Soybeans having high germination rates and ultra-low raffinose and stachyose content. US Patent US20110003045 A1
Sebastian SA, Kerr PS, Pearlstein RW, Hitz WD (2000) Soybean germplasm with novel genes for improved digestibility. In: Drackley JK (ed) Soy in animal nutrition, Federation of Animal Science Societies, Savoy, pp 56–74
Skoneczka JA, Maroof MAS, Shang C, Buss GR (2009) Identification of candidate gene mutation associated with low stachyose phenotype in soybean line PI200508. Crop Sci 49:247
Van den Ende W (2013) Multifunctional fructans and raffinose family oligosaccharides. Front Plant Sci 4:247
Vuong TD, Sleper DA, Shannon JG, Nguyen HT (2010) Novel quantitative trait loci for broad-based resistance to soybean cyst nematode (Heterodera glycines Ichinohe) in soybean PI 567516C. Theor Appl Genet 121:1253–1266
Wu X, Ren C, Joshi T, Vuong T, Xu D, Nguyen HT (2010) SNP discovery by high-throughput sequencing in soybean. BMC Genom 11:469
Zhou ML, Zhang Q, Zhou M, Sun ZM, Zhu XM, Shao JR, Tang YX, Wu YM (2012) Genome-wide identification of genes involved in raffinose metabolism in Maize. Glycobiology 22:1775–1785
Zuther E, Buchel K, Hundertmark M, Stitt M, Hincha DK, Heyer AG (2004) The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett 576:169–173
Acknowledgments
This research was supported by the United Soybean Board, Missouri Soybean Merchandizing Council, and National Center for Soybean Biotechnology (USDA-CSREES Award No. 2008-34555-19305). The authors acknowledge Dennis Colin Yungbluth, Theresa Musket, Division of Plant Sciences, University of Missouri, for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors have no conflict of interest.
Ethical standards
The experiments were performed in compliance with the current laws of the USA.
Additional information
Communicated by V. Hahn.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Qiu, D., Vuong, T., Valliyodan, B. et al. Identification and characterization of a stachyose synthase gene controlling reduced stachyose content in soybean. Theor Appl Genet 128, 2167–2176 (2015). https://doi.org/10.1007/s00122-015-2575-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2575-0