Abstract
Key message
Markers closely flanking a Type 1 FHB resistance have been produced and the potential of combining this with Type 2 resistances to improve control of FHB has been demonstrated.
Abstract
Two categories of resistance to Fusarium head blight (FHB) in wheat are generally recognised: resistance to initial infection (Type 1) and resistance to spread within the head (Type 2). While numerous sources of Type 2 resistance have been reported, relatively fewer Type 1 resistances have been characterised. Previous study identified a Type 1 FHB resistance (QFhs.jic-4AS) on chromosome 4A in Triticum macha. Little is known about the effect of combining Type 1 and Type 2 resistances on overall FHB symptoms or accumulation of the mycotoxin deoxynivalenol (DON). QFhs.jic-4AS was combined independently with two Type 2 FHB resistances (Fhb1 and one associated with the 1BL/1RS translocation). While combining Type 1 and Type 2 resistances generally reduced visual symptom development, the effect on DON accumulation was marginal. A lack of polymorphic markers and a limited number of recombinants had originally prevented accurate mapping of the QFhs.jic-4AS resistance. Using an array of recently produced markers in combination with new populations, the position of QFhs.jic-4AS has been determined to allow this resistance to be followed in breeding programmes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fusarium head blight (FHB) of wheat is caused by several fungal species. Fusarium graminearum is the major pathogen worldwide, but F. culmorum tends to predominate in maritime regions, and F. avenaceum and F. poae are also frequently associated with FHB in Northern Europe. Although FHB may cause large reductions in grain yield and baking quality, a greater threat is posed by mycotoxins which contaminate infected grain and pose a risk to human and animal health. F. graminearum and F. culmorum both produce trichothecene mycotoxins such as deoxynivalenol (DON) and nivalenol (NIV). In addition to the true Fusarium species, two Microdochium species, M. majus and M. nivale, also cause FHB and are particularly prevalent where cooler, wetter conditions prevail, such as the UK. In contrast to F. graminearum and F. culmorum, neither Microdochium species are known to produce mycotoxins, but they may still cause significant reductions in grain quality and yield, particularly in cool wet summers when it may outcompete the Fusarium species (Nicholson et al. 2003). Both the non-toxin producing Microdochium species and the toxin producing Fusarium species can initially infect and colonise wheat spikelets, however, only Fusarium species will spread through the rachis to infect adjacent spikelets. Additionally, it has been shown that although F. graminearum mutants that do not produce DON can colonise wheat heads, they are not able to spread from the point of infection through the rachis (Bai et al. 2002). This suggests that DON is not required for initial infection but is a virulence factor that is necessary for disease spread through the head.
Crop management and agrochemical measures are only partly effective in controlling the disease and therefore the development of FHB resistant varieties is important for disease control and the prevention of mycotoxin contamination. Resistances have been identified in a variety of sources including from Asia (e.g. Sumai-3) (Bai et al. 1999; Waldron et al. 1999), South America (e.g. Frontana) (Schroeder and Christensen 1963; Steiner et al. 2004) and Europe (e.g. Arina) (Snijders 1990). Inheritance of resistance to FHB in wheat is quantitative with a large volume of literature identifying more than 100 quantitative trait loci (QTL) for resistance (Buerstmayr et al. 2009). Several forms of resistance to FHB have been postulated but resistance is generally differentiated into two types: Type 1 (resistance to initial infection) and Type 2 (resistance to spread within the head) (Schroeder and Christensen 1963). The majority of resistance QTL identified confer type 2 resistance (Buerstmayr et al. 2009). This includes the potent 3BS QTL derived from Sumai-3, Qfhs.ndsu-3BS (Anderson et al. 2001), which was subsequently mapped as a single Mendelian gene termed Fhb1 (Cuthbert et al. 2006), and a QTL identified on chromosome 1B that is thought to be located on or closely linked to the 1BL-1RS wheat-rye translocation (Ittu et al. 2000; Shen et al. 2003; Schmolke et al. 2005). Type 1 resistance is considered to be advantageous, because it confers resistance to colonisation both by toxin producing Fusarium species and non-toxin producing Microdochium species. However, it is difficult to identify and select for Type 1 resistance as the presence of this form of resistance must be inferred following assessment by both single spikelet (point) inoculation to assess Type 2 resistance and spray inoculation to assess both Type 1 and Type 2 resistance (Mesterházy et al. 2008). Type 1 resistance QTLs that have been identified include QTL located on chromosome 5A (Buerstmayr et al. 2003; Lin et al. 2006; Steiner et al. 2004), 4B (Lin et al. 2006) and 4A (Steed et al. 2005).
Resistance to FHB has also been described within related species of wheat. In particular, FHB resistance has been identified in T. macha, a hulled hexaploid wheat endemic in the Caucasus region (Dardis and Walsh 2003; Gilbert and Tekauz 2000). FHB resistance QTLs were identified in T. macha on chromosomes 2A, 2B, 5A and 5B using backcross derived recombinant inbred lines (Buerstmayr et al. 2011) and on chromosome 4A using a set of single chromosome substitution lines (Steed et al. 2005). The T. macha 4A resistance was shown to confer Type 1 resistance as it was clearly observed from spray inoculations but was not evident following point inoculation. Importantly, this resistance was shown to reduce both visual disease symptoms and levels of DON, suggesting that it may be useful for deployment in elite varieties to provide protection against FHB. This resistance was mapped as a single gene to 4AS using a double haploid (DH) population, where it co-segregated with the SSR marker Gwm165 and was named QFhs.jic-4AS. However, the limited number of recombinants (43 lines) combined with a lack of polymorphic distal flanking markers prevented accurate localisation of the QTL (Steed et al. 2005).
More than 100 QTL for FHB resistance have been identified and reported in wheat, as reviewed by Buerstmayr et al. (2009). To provide a high level of resistance to FHB in wheat, marker assisted selection (MAS) of these QTL can be used to pyramid these resistances into an agronomically desirable background. This approach relies on the selection of resistances that function additively to confer an enhanced level of resistance when combined together. It is possible that combining Type 1 resistances such as the T. macha 4AS resistance with Type 2 resistances such as the 1B QTL (associated with the 1BL-1RS wheat-rye translocation) and the major 3B QTL (Fhb1), may provide an additive effect restricting both initial infection and subsequent spread of the pathogen along the rachis.
In the present study, we tested combinations of the 1B, 3B and 4A FHB resistance QTL, as outlined above, in a susceptible UK wheat background (Hobbit-‘sib’) to examine if pyramiding FHB resistances will confer additional resistance, both in terms of visual disease symptoms and DON content. In addition, we utilised a 288 line F4 population developed from the susceptible parent Hobbit ‘sib’ and the resistant line DH81, previously developed by Steed et al. (2005), to refine the localisation of the 4AS T. macha Type 1 resistance and to identify SNP markers to aid MAS and pyramiding with other FHB resistance QTL by plant breeders.
Materials and methods
Plant material and population development
Seed of the highly FHB susceptible UK variety Hobbit ‘sib’ (HS) was obtained from the John Innes Centre (JIC) wheat collection, and seed of the highly resistant variety WEK0609® (Gosman et al. 2005) was provided by Pioneer Hi-Bred International Inc. Previous SSR haplotyping has suggested that this variety has a number of QTL providing FHB resistance, including Fhb1 on chromosome 3B and the 1BL-1RS associated resistance (Gosman et al. 2007). Seed was also obtained of the single chromosome substitution line Hobbit ‘sib’/T. macha 4A (HS/Tm4A), and a single chromosome recombinant double haploid line (DH81) previously developed by Steed et al. (2005) from the cross between HS/Tm4A × Hobbit ‘sib’ and shown to possess the FHB QTL.
A single chromosome substitution series was generated for HS × WEK0609® in a Hobbit ‘sib’ background. Single chromosome substitution lines (F6) of chromosomes 1B and 3B were crossed to DH81 and the resulting F2 progeny were screened for the presence/absence of simple sequence repeat (SSR) alleles associated with the 1B, 3B and 4A resistances (see below). This procedure was used to generate a total of 16 independent ‘QTL combination’ lines with a common susceptible background with: the 4A (three lines), 3B (one line) or 1B (two lines) resistance QTL in isolation; a combination of 4A and 1B QTLs (three lines); a combination of 4A and 3B QTLs (four lines); or lacking any FHB QTL (three lines). These ‘QTL combination’ lines were evaluated for FHB resistance in a polytunnel trial and five field trials across 3 years as detailed below.
DH81 was backcrossed to HS and a population of 288 F4 plants was generated. Homozygous recombinant F4 lines (39 lines) identified within this population were selfed (F5) and then bulked for use in phenotypic evaluations of FHB resistance. F4 lines that were recombinant but heterozygous for one or more loci were selfed and the resulting F5 individuals genotyped to identify additional homozygous recombinants (39 lines). Seed of individual plants was then bulked for use in phenotypic evaluations of FHB resistance in three field trials and one polytunnel trial as detailed below.
Simple sequence repeat (SSR) marker analysis
SSR primer sets used were from IPK Gatersleben (Gwm), Wheat Microsatellite Consortium (Wmc), Beltsville Agricultural Research Station (Barc) and INRA (Cfa/Cfd/Gpw), and are described on the GrainGenes website (http://wheat.pw.usda.gov/cgi-bin/graingenes/). PCR conditions were as described by (Bryan et al. 1997).
To identify the presence/absence of the 3B, 1B and 4A QTLs in QTL combination lines, the following SSR probes were used: for 3BS, Gwm389, Gwm533 and Gwm493 (Anderson et al. 2001); for 1B, Barc008 and Gwm018 (Shen et al. 2003); and for 4A, Gwm165, Gwm601 and Gwm610 (Steed et al. 2005).
To identify SSR markers for mapping the T. macha 4A resistance, HS and DH81 were screened with 39 publically available SSR markers that were reported to be located on chromosome 4A, to identify polymorphic and co-dominant markers. Polymorphic SSR markers (Table S1) were applied to the HS × DH81 F4 population and the resulting F5 recombinant lines. DNA extractions, PCR conditions and product size determination were conducted as described by Burt et al. (2011).
Single nucleotide polymorphism (SNP) analysis
The parent lines of the population (HS and DH81) and the single chromosome substitution line HS/Tm4A were screened at LGC Genomics (www.lgcgenomics.com) with a wheat SNP panel using their proprietary KBioscience Competitive Allele-Specific Polymerase chain reaction (KASP) genotyping technique. This SNP panel was developed in conjunction with the University of Bristol and contains over 5000 validated SNP assays (Allen et al. 2013). Polymorphic markers were identified to provide an even coverage of chromosome 4AS and primer sets obtained (Table S1) to apply to the HS × DH81 F4 population and the resulting F5 recombinant lines.
To identify additional 4AS polymorphisms, the parent lines and HS/Tm4A were run on the iSelect 90 k wheat SNP chip (Wang et al. 2014) at the University of Bristol Genomics Facility (http://www.bristol.ac.uk/biology/research/transcriptomics/). The analysis of alleles was conducted using Genome Studio Data Analysis Software from Illumina (http://www.illumina.com/informatics/sequencing-microarray-data-analysis/genomestudio.ilmn) with a cluster file created by Wang et al. (2014) that was trained on a diversity panel of wheat landraces. The sequence of the polymorphic markers was aligned to the flow-sorted scaffolds from the International Wheat Genome Sequencing Consortium (IWGSC) chromosome survey sequence, [available from EnsemblPlants, release 21 (Kersey et al. 2012)] using BLAT (Kent 2002). A BioRuby script (Goto et al. 2010) was used to select the alignment with the highest score and used to infer the likely chromosomal location. Selected iSelect SNPs on 4AS and other chromosomes containing T. macha introgressions were converted into KASP assays by identifying homoeologue SNPs from the survey sequence data and using these in conjunction with the varietal SNPs to design homoeologue specific KASP assays.
QTL combination lines were additionally screened for the presence of the 3B QTL using an Fhb1 linked KASP assay wMAS000008 to confirm the presence of this resistance as determined by SSR markers. This marker was previously developed by Gina Brown-Guedira (USDA) as part of a panel of KASPs for MAS of agronomically important genes in wheat (http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/kasp_download.php?URL).
DNA was extracted from all samples as described by Burt et al. (2011), quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific) and diluted to 10 ng/ul in sterile distilled water for use in KASP–SNP PCR reactions. An 8.112 µl reaction volume consisted of 4 µl of DNA, 4 µl KASP reaction mix (LGC Genomics), and 0.112 µl assay mix (containing 12 µM each allele-specific forward primer and 30 µM reverse primer). The following PCR conditions were used: 15 min at 94 °C; 10 touchdown cycles of 20 s at 94 °C, 60 s at 65–57 °C (decreasing 0.8 °C per cycle); and 26–35 cycles of 20 s at 94 °C, 60 s at 57 °C. Fluorescence detection of the reactions was performed using a Bio-Rad CFX96 real-time PCR machine to conduct end point allelic discrimination with CFX Manager 3.1 software (Bio-Rad Laboratories).
Conserved orthologous sequence (COS), expressed sequence tag SSR (EST-SSR) and SSR marker analysis
Sequences for the polymorphic wheat genotyping panel SNPs and the polymorphic iSelect SNPs were aligned to the Brachypodium, rice and Sorghum genomes using Phytozome v9.1 (www.phytozome.net) to identify the orthologous loci in these species, where present. The Brachypodium sequence corresponding to the region of interest on wheat 4AS was visually examined using the Brachypodium (Bd21) Genome Browser (http://www.modelcrop.org/cgi-bin/gbrowse/brachyv1/) to identify thirty-three COS markers aligned to the region. These markers were previously developed by Dr. Simon Griffiths and Michelle Leverington-Waite at the John Innes Centre.
Expressed sequence tag-derived microsatellite (EST-SSRs) for comparative mapping in wheat, barley and rice were previously identified by La Rota et al. (2005). From these, primers for a set of 26 EST-SSRs were identified in the rice region orthologous to 4AS in wheat. The 33 COS and 26 EST-SSR markers were tested on the parent lines (HS and DH81) to identify polymorphisms on 4AS located in the region of the resistance. Polymorphic markers (Table S1) were applied to the HS × DH81 F4 population and the resulting F5 recombinant lines.
DNA extractions and PCR reactions were prepared as described by Burt et al. (2011). PCR amplification was conducted using a touchdown programme consisting of a denaturing step of 95 °C for 10 min; 16 touchdown cycles of 95 °C for 15 s, 58 °C for 1 min decreasing 0.5 °C per cycle, 72 °C for 1 min; then 30 cycles of 95 °C for 15 s, 50 °C for 15 s and 72 °C for 1 min. Samples were run on an ABI 3700 capillary sequencer (Applied Biosystems) and the output data were analysed using Peak Scanner v1.0 (Applied Biosystems) to determine the product size of the amplicons. If no polymorphism was detected using this method, then products were examined by single-strand conformation polymorphism (SSCP) assay (Martins-Lopes et al. 2001) using Sequa GelMD (National Diagnostics, UK Ltd.) and visualised by silver staining (Bassam et al. 1991).
FHB resistance phenotyping of the 1B, 3B and 4A QTL combination lines
16 lines representing the 5 QTL combination categories [1B, 3B, 1B and 4A, 3B and 4A, and Null (none)] were entered into five independent field trials across 2 years. These were: John Innes Centre, Norfolk, UK (JIC) and Tulln, Austria in 2011; and JIC, Tulln, and Church Farm, Bawburgh, Norfolk, UK (CF) in 2012.
These field experiments were conducted in a randomised complete block design with four and two replicate blocks per line in the UK and Austria, respectively. Trials in the UK were inoculated with a highly virulent DON-producing F. culmorum isolate (Fu42), whilst in the trials in Austria, a highly aggressive F. graminearum isolate (IFA66) was used. In the UK, plants were spray inoculated with a conidial suspension (1 × 104 ml−1) amended with 0.05 % Tween20 at mid anthesis [growth stage (GS) 65, (Zadoks et al. 1974)] using a knapsack sprayer (150 ml m−2). Plants were mist irrigated for a minimum of 72 h post inoculation to maintain high humidity. The inoculation was repeated after an interval of 3 days. Disease was assessed as % infection within each plot at four time intervals post infection (dpi) to follow disease development in each trial. The area under the disease progress curve (AUDPC) was calculated from percentage infection at each time point to provide an integrated measure of disease.
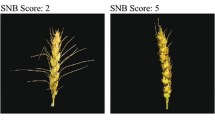
Additionally, an experiment was conducted in an unheated polytunnel at JIC in 2010 with 4 replicate plants per line arranged in a randomised block design on capillary matting. Inoculum was prepared as described above and plants were inoculated at GS65 with a conidial suspension (1 × 104 ml−1) amended with 0.05 % Tween20 at GS65 using a held–held sprayer. Following spray inoculation, plants were visually assessed for disease on the basis of percentage of spikelets infected per head at 10, 14 and 18 dpi (%FHB). Percentage spikelet infection at 18 dpi was used as a measure of disease severity. The area under the disease progress curve (AUDPC) was again calculated to provide an integrated measure of disease development.
In Austria, all the plots (2 rows, 80 cm long) were repeatedly sprayed with 100 ml m−2 of a 1 × 104 conidia/ml macroconidial suspension using a backpack sprayer. The first inoculation was done 2 days before the earliest line flowered and the treatment was repeated (in total 6 applications with 2 day time intervals) until the last line reached full anthesis. During and after each inoculation cycle, the crop canopy was kept wet with a computer controlled mist irrigation system for 20 h. GS 65 was assessed for each line individually. Ten days after mid anthesis, visual disease assessment started and was repeated on day 14, 18, 22 and 26 after mid anthesis. The percentage of visually diseased spikelets was recorded with which the AUDPC was calculated.
Grain samples were taken from the JIC and Tulln field trials in both 2011 and 2012, and the polytunnel trial in 2010. Samples were milled and DON was extracted in 10 % methanol and DON content was assessed using the Ridascreen Fast DON™ (R-Biopharm Rhone Ltd.) enzyme linked immuno-assay (ELISA) according to the manufacturer’s instructions as described previously (Gosman et al. 2005).
FHB resistance phenotyping of the HS × DH81 population
In total, 78 recombinant lines were selected from the HS × DH81 F4 population for use in the current study. Thirty nine stable recombinant F5 lines initially identified and generated from the F4 population were assessed for FHB resistance in a field trial at JIC in 2012. The FHB resistance of all 78 recombinant F5 lines was assessed during the summer of 2013 in two independent field trials; one at CF and one at JIC. These field experiments were conducted in a randomised complete block design with three replicate plots per line. All trials were inoculated with a highly virulent DON-producing F. culmorum isolate (Fu42) and conducted as described above. Disease was assessed as % infection within each plot at 16, 22, 25 and 30 days post infection (dpi). The area under the disease progress curve (AUDPC) was again calculated to provide an integrated measure of disease and % infection at 30 dpi was used as a measure of disease severity (%FHB).
The 39 stable recombinant HS × DH81 F5 lines initially identified and generated from the F4 population were assessed for FHB resistance in 2013 at JIC in an unheated polytunnel with capillary matting irrigation. Fifteen plants per line were arranged in a randomised complete block design with 4 blocks (3–4 plants per line within each block). Inoculations were conducted and plants were scored as described for the above polytunnel trial.
Statistical analysis
Analysis of variance (ANOVA) using a general linear model (GLM) was performed on AUDPC scores and DON contents (ppm) from the independent FHB resistance experiments for the 1B, 3B and 4A QTL combination lines to assess variation due to QTL class and experiment × QTL class interaction. Means were predicted across the relevant lines for each QTL class, and were subsequently compared using Fisher’s least significant difference test.
ANOVA using a GLM was also performed on %FHB and AUDPC scores from the four phenotyping experiments for the HS × DH81 population to assess variation due to line and experiment × line interaction. This was conducted separately for 39 line experiments (using all data: 2012 JIC, 2013 JIC, 2013 CF and 2013 Polytunnel) and for the 78 line experiments (2013 JIC and 2013 CF) to provide balanced datasets for analysis. Predicted mean disease scores were calculated for the lines within the GLMs. In addition, GLMs were fitted for each experiment individually and predicted mean scores for each line calculated within the models to account for variation due to field block. Means calculated across experiments and means calculated within experiments were used in subsequent QTL analyses.
All GLMs were conducted in Genstat v. 15.2. Broad sense heritability across experiments was estimated from the ANOVA using the formula: \(H = \, {{\sigma_{\text{G}}^{{^{ 2} }} } \mathord{\left/ {\vphantom {{\sigma_{\text{G}}^{{^{ 2} }} } {\left[ {\sigma_{\text{G}}^{{^{ 2} }} + \, \left( {\sigma_{\text{GE}}^{{^{ 2} }} /{\text{E}}} \right) \, + \, \left( {\sigma_{\text{e}}^{{^{ 2} }} /{\text{rE}}} \right)} \right]}}} \right. \kern-0pt} {\left[ {\sigma_{\text{G}}^{{^{ 2} }} + \, \left( {\sigma_{\text{GE}}^{{^{ 2} }} /{\text{E}}} \right) \, + \, \left( {\sigma_{\text{e}}^{{^{ 2} }} /{\text{rE}}} \right)} \right]}}\), with \(\sigma_{\text{G}}^{{^{ 2} }}\) the genetic variance, \(\sigma_{\text{GE}}^{{^{ 2} }}\) the genotype × experiment interaction variance, \(\sigma_{\text{e}}^{{^{ 2} }}\) the residual variance, E the number of experiments, and r the number of replicates per genotype (Nyquist 1991).
Map construction and QTL analysis
A genetic linkage map of chromosome 4A was constructed using 14 LGC KASP wheat SNPs, 2 iSelect SNP derived KASPs, 3 SSRs and 2 EST-SSRs applied to the DNA from 288 F4 lines. The linkage analysis was performed in Joinmap (version 3.0) (Van Ooijen and Voorips 2001), using 0.4 as the maximum recombination fraction and 5.0 as the logarithm of the odds ratio (LOD), and the linkage map was drawn using MapChart (Voorrips 2002).
Predicted mean AUDPC and %FHB scores were calculated for the 39 HS × DH81 F5 recombinant lines across all 4 experiments (2012 JIC, 2013 JIC, 2013 CF and 2013 Polytunnel), and for all of the 78 lines HS × DH81 F5 recombinant lines across the two experiments in which all lines were included (2013 JIC and 2013 CF). These predicted means were then used alongside marker data from the same lines in simple interval mapping (SIM) in Genstat v.15.2. Significance of QTL were presented as p values on a log10 scale [−log10(p)]. To declare QTL, a −log10(p) threshold of 2.342 was determined using the method of Li and Ji (2005) with a genome wide significance level of 0.05.
To check for stability of QTL effects across different trials, predicted mean %FHB and AUDPC scores from the field trials and polytunnel experiment of F5 recombinant lines were used alongside marker data from the same lines in a single marker regression analysis to identify QTL locations for each trait within each experiment. A single marker regression analysis was utilised as there were relatively few markers (21) densely spaced on a single linkage group. Markers were only determined to be associated with the phenotype, where p < 0.01, to reduce the likelihood of false positives.
Results
FHB resistance in the 1B, 3B and 4A QTL combination lines
SSR haplotype data identified 16 lines with the following combinations of QTL: (i) the 3B QTL alone (1 line), (ii) the 1B QTL alone (2 lines), (iii) the 4A QTL alone (3 lines), (iv) 4A and 3B QTLs (4 lines), (v) 4A and 1B QTLs (3 lines), and (vi) ‘Null’ lines (3 lines) as susceptible controls that had been through the crossing procedure but lacked any of the QTL according to SSR haplotype data. The presence or absence of the 3B QTL in the QTL combination lines as predicted by the SSR data was additionally confirmed by wMAS000008.
Data were combined across the lines for each QTL class and also across the trials to conduct an analysis of variance and to predict means for AUDPC and DON contents. Effects of the QTL combination classes were significant for both AUDPC and DON contents (Table 1). However, there was a significant class by trial interaction for both AUDPC and DON contents (p < 0.001)) suggesting that there were differences in the relative performance of the lines across the trials. Broad sense heritability estimates were relatively high for AUDPC (0.80) suggesting a high level of stability of the effect of the QTL combinations in different environments. However, DON contents were less stable across environments for the QTL combinations with a heritability of 0.60 (Table 1).
Predicted means for AUDPC and DON contents demonstrated that the 1B, 3B and 4A QTL individually confer a significant reduction (p < 0.05) in both visual disease scores and DON levels compared to the ‘Null’ lines without any known resistance QTL (Fig. 1). There was some evidence that combinations of QTLs may have an additive effect on visual disease scores. The combination of 4A and 1B FHB QTLs conferred a significant reduction in visual disease scores compared to either 4A alone or 1B alone (p < 0.05) (Fig. 1a). Although the 3B and 4A combined resistance provided some evidence of enhanced disease control compared to the 4A QTL alone, the reduction was not significant (Fig. 1a).
Predicted mean a AUDPC scores b DON contents of grain samples in ppm for the QTL classes calculated across trials. Error bars are all ± standard error of the mean. Predicted are compared using Fisher’s least significant difference test and comparisons demonstrating significant differences (p < 0.05) are shown as: a from Null; b from 1B; c from 3B; d from 4A
There was no evidence that combining QTL enhances resistance to DON accumulation. Combining the 1B and 4AQTL reduced the amount of DON compared to either QTL in isolation (Fig. 1b), however, these reductions were not significant (p > 0.05). The combination of 4A and 3B QTLs provided a similar level of reduction in DON content compared to the 3B QTL alone.
Differences in FHB resistance are often associated with plant height (Srinivasachary et al. 2008). All lines in the current work, however, were of similar height so removing this aspect for consideration.
HS × DH81 marker analysis, genotyping and map construction
Of the 115 LGC wheat KASP–SNP markers previously shown to be on chromosome 4AS (Allen et al. 2011, 2013), 20 were polymorphic (Table S2). From these, a sub-set of 14 co-dominant and polymorphic SNPs was identified to provide an even coverage of chromosome 4AS on the basis of published SNP maps of the Avalon × Cadenza and Savannah × Rialto populations (Allen et al. 2011, 2013). These 14 SNP markers were applied to the HS × DH81 F4 population and also to the resulting F5 recombinant lines to confirm the genotypes. The 33 COS and 26 EST-SSR markers were tested on HS and DH81 to identify polymorphisms. Only two EST-SSR markers were polymorphic and these were applied to the HS × DH81 F3 population and F4 recombinant lines. No COS markers were polymorphic and were therefore not applied to the population (Table S2). Of the 39 SSR markers tested, 9 were polymorphic of which 3 were co-dominant (Wmc48, Gwm192 and Gwm165). One of these, Gwm165, was previously identified by Steed et al. (2005). These three SSR markers were applied to the HS × DH81 F4 population and subsequent F5 recombinant lines.
Eighty-three polymorphic iSelect markers were identified on 4AS. From these, 6 markers were identified by BlastN to have homology to Brachypodium, rice and Sorghum genes within the region orthologous to the QTL location as initially located by the LGC KASP, SSR and EST-SSR markers (Fig. 2). Two markers on 4AS were successfully converted to co-dominant KASP assays (BS00182960 and BS00164805) and were applied to the HS × DH81 F4 and F5 lines.
Linkage map of chromosome 4AS of Hobbit ‘sib’ (HS) × DH81 compared to the physical marker locations on Brachypodium chromosome 1, Sorghum chromosome 1 and rice chromosome 3. Genetic map distances in HS × DH81 are indicated in cM on the scale on the left. The shaded area indicates the approximate region of the 4A QTL
In total, 14 LGC wheat KASP SNPs, 2 iSelect derived KASPs, 3 SSRs and 2 EST-SSR markers (Fig. 2) were applied to the 288 lines in the HS × DH81 F4 population to construct a genetic map totalling 70.6 cM (Fig. 2). These 21 markers were also applied to each of the 78 recombinant F5 lines to confirm the genotypes.
Regions in Brachypodium, rice and sorghum were identified with synteny to the QFhs.jic-4AS region in wheat from BS00022015 to BS00164805 (Fig. 2). Although within this region, there was a high level of gene order conservation between the three reference genomes; this was not completely conserved in the genetic map order from HS × DH81, with the marker pairs BS00164805 and TC90601, TC93568 and BS00113963, and BS00022015and BS00003776 inverted compared to their orthologues. It was not possible to establish co-linearity outside of this region, with only two wheat markers identifying orthologues (BS00022816 orthologous to Bradi1g11550, BS00003914 orthologous to Bradi4g20120).
HS × DH81 trait analysis
Line effects were significant for both AUDPC and % FHB traits in both 39 and 78 line experiment sets (Table 2). However, there was a significant line by experiment interaction for AUDPC in the 78 line experiments (p < 0.001) suggesting that there were differences in the relative performance of the lines across the two trials. Broad sense heritability for both AUDPC and %FHB traits were higher in the 39 line experiments (Table 2). This may reflect a better estimate of the genetic effects when using 4 environments, and additionally, may be influenced by a greater level of environmental control and more detailed scoring of disease in the polytunnel experiment of 39 lines.
Predicted means of AUDPC and %FHB for the HS × DH81 recombinant F5 lines were plotted in frequency histograms for all experiments (Figure S1). It was not possible to detect a bimodal distribution as identified for the T. macha 4A resistance in Steed et al. (2005), with all experiments providing an approximate normal distribution of means.
HS × DH81 QTL analysis
Significant QTL originating from DH81 and conferring FHB resistance was detected in both the 39 line set and 78 line sets. The peak QTL position was located on the marker BS00011173 for AUDPC in both sets of lines and the same marker was identified as the peak QTL position for %FHB in the 78 line set. A slightly different location at TC93568 was identified as the QTL peak for %FHB in the 39 line set. However, this marker is only 3.1 cM away (Table 3), suggesting that this represents the same genetic effect. The QTL scan shows clear QTL peaks, particularly for %FHB across all 78 lines. It also shows consistent QTL location with LOD scores above the significant threshold for all 4 QTL analyses in the region approximately between 30 and 40 cM (Fig. 3). In addition to the main QTL identified at BS00011173 and TC93568, additional resistance QTL originating from HS were detected in the 39 line set at marker Wmc48 for %FHB and at marker BS00003623 for AUDPC.
A single marker regression of individual trial locations identified that the major resistance QTL from DH81 identified in the SIM analysis was consistent across the experiments (Table 4). For AUDPC the single marker regression identified a significant association (p < 0.01) between resistance and markers within 3.5 cM of BS00011173 in all trials. Significant associations with %FHB were detected in a region overlapping BS00011173 and TC93568 in three experiments (2013 JIC, 2013 CF and Polytunnel 2013). However, the experiment conducted in 2012 at JIC was less consistent, identifying a QTL at Gwm165, which is only 0.4 cM from TC93568, but also identifying a QTL at BS0006885 and BS00022015, which are 5.8 and 7 cM, respectively, from BS0001173 (Table 4).
Although a QTL from HS conferring a reduction in %FHB was detected at Wmc48, and a QTL from HS conferring a reduction in AUDPC was detected at BS0003623 in the SIM analysis of the 39 line set, the single marker regression of data from individual trials only identified significant associations between this region and disease traits at markers Wmc48 and BS00036472 in the AUDPC trait from the 2013 JIC trial (Table 4). This suggests that this genetic effect is not consistent across environments.
The polytunnel trial found the EST-SSR marker TC93568 accounts for the highest proportion of variation for both AUDPC and %FHB traits, using single marker regression. The greater amount of variation accounted for by markers within the polytunnel trial, compared to the field trials, may be due to a more homogenous environment and/or the more detailed scoring of individual spikelets in this procedure. Steed et al. (2005) conducted all phenotyping in a polytunnel, and this may have assisted the detection of the resistance as a single gene with Mendelian inheritance.
Background effects in the HS × DH81 population
The wheat KASP panel and the wheat iSelect chip detected the presence of polymorphisms between HS and DH81 on chromosomes 4B and 7A (Table S3). These are likely to be due to remaining T. macha introgressions in DH81 that have not been removed by backcrossing. To test if these regions were influencing the phenotype, primers were obtained for the polymorphic wheat SNP panel KASP assays on 4B and 7A. On 4B, the wheat SNP BS00022576 provided a clear assay and was applied to the HS × DH81 F5 recombinants. None of the 7A wheat KASP assays provided clear polymorphisms when tested on the population and therefore the iSelect SNP BS00160015 on 7A was converted into a KASP assay that was also applied to the HS × DH81 F5 recombinants. Single marker regressions to compare these two markers to AUDPC and %FHB in the three trials did not identify any significant relationships (R 2 = 0–7.4 %, p > 0.05), suggesting that these regions are not influencing the observed phenotypes (Table S3). As for the QTL lines, no difference in plant height were observed within the HS × DH81 population.
Discussion
Previous genetic studies have suggested that relatively small effect FHB resistances may function additively to confer a higher level of resistance (Anderson et al. 2001; Buerstmayr et al. 2003; Snijders 1990). We combined the Type 2 1B and 3B resistances with the type 1 resistance from T. macha 4A (QFhs.jic-4AS) in a winter wheat background to test for additive effects when combining these resistances. The recurrent parent used in this experiment was Hobbit ‘sib’, a UK winter wheat with a high level of susceptibility. In particular, it contains Rht-D1b, which has been shown to be highly associated with susceptibility to FHB in numerous studies (Kollers et al. 2013; Srinivasachary et al. 2008, 2009). Despite the high level of susceptibility in the recurrent parent and the high level of disease pressure applied in inoculated, irrigated trials, the 3 resistances all conferred a high level of resistance when deployed individually, reducing both visual disease symptoms and DON content. Both 4A-3B and 4A-1B combinations demonstrated enhanced resistance in terms of reduced visual disease symptoms compared to the individual QTL suggesting that these resistances may function additively to reduce disease. This may be due to the combination of the Type 1 resistance QFhs.jic-4AS, with the Type 2 resistances Fhb1 (3B) and the 1B QTL. However, there was no evidence that these combined resistances functioned additively to reduce the amount of DON that was present in wheat grain at harvest. It is possible that more than one type 2 resistance, which act to prevent DON mediated disease spread, are required to provide an additive reduction in DON levels.
The development of varieties with pyramided FHB resistances will be facilitated by increased mapping accuracy and more markers for selection of resistances. Fhb1 on chromosome 3B and the 1B QTL have been extensively mapped and a number of molecular markers are available for their selection through MAS. However, prior to this study, the map location of QFhs.jic-4AS was imprecise. Previous efforts to map this resistance have been restricted by a lack of polymorphic markers. Steed et al. (2005) utilised existing SSR and developed novel sequence-specific amplified polymorphism (SSAP) markers, but were not able to identify any distal markers to flank the resistance to facilitate marker assisted selection of the resistance by plant breeders. Developments in SNP technology and the availability of wheat SNPs both through the KASP assays (Allen et al. 2013) and the wheat 90 K iSelect genotyping assay (Wang et al. 2014) enabled saturation of the region surrounding the 4AS QTL. It was therefore possible to identify breeder-friendly KASP markers underlying the QTL region such as BS00011173 and BS00113963. It was also possible to identify distal flanking KASP assay markers such as BS0006885 and BS00022015, and proximal flanking markers such as the iSelect derived KASP BS00164805 and the KASP assay BS00036472 that would be suitable for selection of the region containing QFhs.jic-4AS. Although the effect of QFhs.jic-4AS was not potent in trials involving the recombinant lines, it was clear in the QTL combination lines, both alone and in combination with the Type 2 resistances from 1B and 3B.
Previously, Steed et al. (2005) located the QFhs.jic-4AS as a single gene using visual disease symptoms observed in a polytunnel using a population 43 DH lines. The genetic effect of the region as a whole appears to be relatively large, providing heritability estimates of 0.55–0.82 for the FHB resistance traits recorded (Table 3), and the effect of the QTL was highly significant when studied in the QTL combination lines (Fig. 1) In contrast, in the present study using 78 F5 lines from a recombinant population, we were unable to resolve the resistance as a single gene in any experiment or across experiments. However, we were able to locate the resistance quantitatively using SIM and single marker regression, and demonstrate that the effect is consistently identified across the experiments. It is possible that the T. macha 4A resistance is conferred by multiple genes of small effect distributed over the approximately 12.2 cM region between markers BS00011060 and BS00164805. The additional recombinants within the 288 F4 lines and the high marker density in the present study, compared to the limited recombinants within the 43 DH lines studied by Steed et al. (2005), and in the 4A lines studied in the QTL combination field trials in the present study, may have fractionated QFhs.jic-4AS into multiple QTL within a small region. There is some evidence of this from the results obtained from the polytunnel trial when assessing disease severity, which identified three QTL peaks at BS00182960, TC93568 and Gwm192, and also from the JIC trial in 2012, which indicates two QTL conferring resistance to %FHB at Gwm165 and also at BS00022015-BS00068885. However, failure to resolve the QTL as a single gene may reflect the difficulty of accurately phenotyping FHB using only one score. This possibility is evidenced by the fact that the single-score data support a fractionated QTL while the integrated AUDPC score suggests a single QTL centring on marker TC93568. However, from the current data, it is not possible to determine whether there are multiple QTL or whether different loci have been detected as a consequence of unexplained variation in individual experiments. The generation of further recombinants and more detailed disease phenotyping of the lines using the greater accuracy that can be achieved in polytunnel trials are required to determine whether the phenotype observed from QFhs.jic-4AS is conferred by the additive effect of multiple QTL. Several previous studies have identified large effect QTLs that fractionate into multiple linked QTL when fine-mapping. This includes resistance against Phytophthora infestans in tomato (Johnson et al. 2012), the maize domestication QTL teosinte branched 1 (Studer and Doebley 2011), and a malting quality QTL complex in barley (Gao et al. 2004). Other factors may have hindered resolution of QFsh.jic-4AS as a single gene. Accurate phenotyping of Type 1 resistance is recognised to be challenging because of confounding effects of Type 2 susceptibility in lines such as Hobbit sib. used in the present study. Furthermore, disease pressure was extremely high in all trials, as revealed by the high levels of DON, and this may have resulted in the fungus overcoming the resistance conferred by QFsh.jic-4AS. Additional, detailed phenotyping using reduced disease pressure may assist in resolving this issue.
The QFhs.jic-4AS was detected in approximately the same region, using both disease development (represented by AUDPC) and disease severity (represented by %FHB) measurements, in four independent phenotyping experiments. This suggests that the resistance can be considered to be stable, as it was expressed across different environments. Although previously identified as a Type 1 resistance, with no effect on disease spread following point inoculation, this data suggests that this resistance may have additional effects other than limiting initial colonisation (Type 1 resistance) as the resistance effect can be clearly observed when studying disease development over time (AUDPC). In addition, although the effects of the QFhs.jic-4AS QTL can be observed at earlier scoring times (data not shown), it appeared to have greatest effect when scoring visual symptoms after a relatively long period after inoculation (29–30 dpi) supporting the view that it also functions after initial infection.
There are some discrepancies between the trials that should be considered when choosing markers for selection of the QFhs.jic-4AS resistance. For example, BS00164805 would appear to be suitable as a proximal flanking marker based on the AUDPC data from all four trials and from the %FHB data from JIC and the polytunnel trial, providing no evidence of association with the resistance (p < 0.01) and hence suggesting that this marker is located immediately proximal to the resistance. However, BS00164805 provides a significant association with the %FHB scores generated from the CF field experiment, suggesting that this marker may be associated with the QTL based on this analysis. In addition, the % variance accounted for tends to increase for this marker compared to the proximal marker TC90601. It is possible that this marker has been positioned incorrectly by genetic mapping and that it should sit between TC90601 and Gwm192, which would be supported by synteny in the region (Fig. 2). However, it is also possible that this marker is providing further evidence for fractionation of QFhs.jic-4AS.
EST-derived SSRs (La Rota et al. 2005) and wheat KASP assay SNPs derived from transcript sequencing (Allen et al. 2011, 2013) were used to identify orthologues in syntenic regions within the fully sequenced genomes of Brachypodium, rice and sorghum. This enabled the identification of the region in these species that was orthologous to QFhs.jic-4AS. This region was then examined to identify further wheat iSelect SNPs based on their homology with these reference genomes. However, there is some evidence of a breakdown of synteny between the order of loci as determined by the wheat × T. macha genetic map of 4A developed in this study and the physical gene orders in the reference sequences (Fig. 2). A breakdown of co-linearity may be anticipated on chromosome 4A because it has undergone a significant number of rearrangements compared to the structure of related species. Previous studies have identified a peri-centromeric inversion involving a portion of the ancient long arm and the complete short arm, and interchanges with chromosomes 5A and 7B (Devos et al. 1995; Miftahudin et al. 2004). More recently, next-generation sequencing and synteny with Brachypodium, rice and sorghum was used to construct a chromosome 4A ‘genome-zipper’ with five syntenic segments (Hernandez et al. 2012). The QTL region in the present study lies partly within the syntenous chromosomal segment ‘A’ identified by Hernandez et al. (2012) from Bradi1g65190 to Bradi1g72092. The breakdown of gene order conservation within the QTL region and our inability to establish any co-linearity outside of the region, suggests that there may be a limitation in the use of synteny for further fine-mapping of QFhs.jic-4AS, particularly as the resistance may be controlled by multiple loci over the region.
In conclusion, we have demonstrated that Type 1 and Type 2 resistances can be combined in a highly susceptible background to provide an additive reduction in visual disease symptoms. However, caution should be exercised as this reduction in visual symptoms, may not be translated into a reduction in mycotoxin levels. To enhance the capabilities for marker assisted selection of the FHB Type 1 resistance QFhs.jic-4AS, we have developed additional recombinants and identified a number of SNP markers suitable for use by plant breeders. However, we were not able to locate and map the resistance as a single gene.
Author contribution statement
CB undertook mapping, genotyping, phenotyping and writing of the manuscript, NG initiated production of QTL materials, AS undertook crossing and field work, ML undertook FHB trials in Tulln, NB and RR-G assisted with marker development, SH undertook mapping population development, and PN conceived and coordinated the work.
References
Allen AM, Barker GLA, Berry ST, Coghill JA, Gwilliam R, Kirby S, Robinson P, Brenchley RC, D’Amore R, McKenzie N, Waite D, Hall A, Bevan M, Hall N, Edwards KJ (2011) Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol J 9:1086–1099
Allen AM, Barker GLA, Wilkinson P, Burridge A, Winfield M, Coghill J, Uauy C, Griffiths S, Jack P, Berry S, Werner P, Melichar JPE, McDougall J, Gwilliam R, Robinson P, Edwards KJ (2013) Discovery and development of exome-based, co-dominant single nucleotide polymorphism markers in hexaploid wheat (Triticum aestivum L.). Plant Biotechnol J 11:279–295
Anderson JA, Stack RW, Liu S, Waldron BL, Fjeld AD, Coyne C, Moreno-Sevilla B, Fetch JM, Song QJ, Cregan PB, Frohberg RC (2001) DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor Appl Genet 102:1164–1168
Bai G, Kolb FL, Shaner G, Domier LL (1999) Amplified fragment length polymorphism markers linked to a major quantitative trait locus controlling scab resistance in wheat. Phytopathology 89:343–348
Bai GH, Desjardins AE, Plattner RD (2002) Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 153:91–98
Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Bryan GJ, Collins AJ, Stephenson P, Orry A, Smith JB, Gale MD (1997) Isolation and characterisation of microsatellites from hexaploid bread wheat. Theor Appl Genet 94:557–563
Buerstmayr H, Steiner B, Hartl L, Griesser M, Angerer N, Lengauer D, Miedaner T, Schneider B, Lemmens M (2003) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor Appl Genet 107:503–508
Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128:1–26
Buerstmayr M, Lemmens M, Steiner B, Buerstmayr H (2011) Advanced backcross QTL mapping of resistance to Fusarium head blight and plant morphological traits in a Triticum macha × T. aestivum population. Theor Appl Genet 123:293–306
Burt C, Hollins TW, Nicholson P (2011) Identification of a QTL conferring seedling and adult plant resistance to eyespot on chromosome 5A of Cappelle Desprez. Theor Appl Genet 122:119–128
Cuthbert P, Somers D, Thomas J, Cloutier S, Brulé-Babel A (2006) Fine mapping Fhb1, a major gene controlling Fusarium head blight resistance in bread wheat (Triticum aestivum L.). Theor Appl Genet 112:1465–1472
Dardis JV, Walsh EJ (2003) An evaluation of exotic germplasm as sources of Fusarium head blight resistance in bread wheat (Triticum aestivum L.). Cereal Research Communications 31:129–136
Devos KM, Dubcovsky J, Dvořák J, Chinoy CN, Gale MD (1995) Structural evolution of wheat chromosomes 4A, 5A, and 7B and its impact on recombination. Theor Appl Genet 91:282–288
Gao W, Clancy JA, Han F, Jones BL, Budde A, Wesenberg DM, Kleinhofs A, Ullrich SE (2004) Fine mapping of a malting-quality QTL complex near the chromosome 4H S telomere in barley. Theor Appl Genet 109:750–760
Gilbert J, Tekauz A (2000) Review: recent developments in research on Fusarium head blight of wheat in Canada. Can J Plant Pathol 22:1–8
Gosman N, Chandler E, Thomsett M, Draeger R, Nicholson P (2005) Analysis of the relationship between parameters of resistance to Fusarium head blight and in vitro tolerance to deoxynivalenol of the winter wheat cultivar WEK0609 ®. Eur J Plant Pathol 111:57–66
Gosman N, Bayles R, Jennings P, Kirby J, Nicholson P (2007) Evaluation and characterization of resistance to Fusarium head blight caused by Fusarium culmorum in UK winter wheat cultivars. Plant Pathol 56:264–276
Gosman N, Steed A, Faure S, Bayles R, Jennings P, Nicholson P (2008) Mapping of QTL associated with Fusarium head blight in spring wheat RL4137. Czech J Genet Plant Breed 44:147–159
Goto N, Prins P, Nakao M, Bonnal R, Aerts J, Katayama T (2010) BioRuby: bioinformatics software for the Ruby programming language. Bioinformatics 26:2617–2619
Hernandez P, Martis M, Dorado G, Pfeifer M, Gálvez S, Schaaf S, Jouve N, Šimková H, Valárik M, Doležel J, Mayer KFX (2012) Next-generation sequencing and syntenic integration of flow-sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content. Plant J 69:377–386
Ittu M, Saulescu NN, Hagima I, Ittu G, Mustatea P (2000) Association of Fusarium head blight resistance with gliadin loci in a winter wheat cross. Crop Sci 40:62–67
Johnson EB, Haggard JE, Stclair DA (2012) Fractionation, stability, and isolate-specificity of QTL for resistance to Phytophthora infestans in cultivated tomato (Solanum lycopersicum). G3: Genes|Genomes|Genetics 2:1145–1159
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Kersey PJ, Staines DM, Lawson D, Kulesha E, Derwent P, Humphrey JC, Hughes DST, Keenan S, Kerhornou A, Koscielny G, Langridge N, McDowall MD, Megy K, Maheswari U, Nuhn M, Paulini M, Pedro H, Toneva I, Wilson D, Yates A, Birney E (2012) Ensembl genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucl Acids Res 40:D91–D97
Kollers S, Rodemann B, Ling J, Korzun V, Ebmeyer E, Argillier O, Hinze M, Plieske J, Kulosa D, Ganal MW, Röder MS (2013) Whole genome association mapping of Fusarium head blight resistance in European winter wheat (Triticum aestivum L.). PLoS ONE 8:e57500
La Rota M, Kantety RV, Yu JK, Sorrells ME (2005) Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC Genom 6:23
Li J, Ji L (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95:221–227
Lin F, Xue SL, Zhang ZZ, Zhang CQ, Kong ZX, Yao GQ, Tian DG, Zhu HL, Li CJ, Cao Y, Wei JB, Luo QY, Ma ZQ (2006) Mapping QTL associated with resistance to Fusarium head blight in the Nanda2419 × Wangshuibai population. II: type I resistance. Theor Appl Genet 112:528–535
Martins-Lopes P, Zhang H, Koebner R (2001) Detection of single nucleotide mutations in wheat using single strand conformation polymorphism gels. Plant Mol Biol Rep 19:159–162
Mesterházy A, Tóth B, Bartók T, Varga M (2008) Breeding strategies against FHB in winter wheat and their relation to type I resistance. Cereal Res Commun 36:37–43
Miftahudin Ross K, Ma X-F, Mahmoud AA, Layton J, Milla MAR, Chikmawati T, Ramalingam J, Feril O, Pathan MS, Momirovic GS, Kim S, Chema K, Fang P, Haule L, Struxness H, Birkes J, Yaghoubian C, Skinner R, McAllister J, Nguyen V, Qi LL, Echalier B, Gill BS, Linkiewicz AM, Dubcovsky J, Akhunov ED, Dvořák J, Dilbirligi M, Gill KS, Peng JH, Lapitan NLV, Bermudez-Kandianis CE, Sorrells ME, Hossain KG, Kalavacharla V, Kianian SF, Lazo GR, Chao S, Anderson OD, Gonzalez-Hernandez J, Conley EJ, Anderson JA, Choi D-W, Fenton RD, Close TJ, McGuire PE, Qualset CO, Nguyen HT, Gustafson JP (2004) Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168:651–663
Nicholson P, Chandler E, Draeger RC, Gosman NE, Simpson DR, Thomsett M, Wilson AH (2003) Molecular tools to study epidemiology and toxicology of Fusarium head blight of cereals. Eur J Plant Pathol 109:691–703
Nyquist WE (1991) Estimation of heritability and prediction of selection response in plant-populations. Crit Rev Plant Sci 10:235–322
Schmolke M, Zimmermann G, Buerstmayr H, Schweizer G, Miedaner T, Korzun V, Ebmeyer E, Hartl L (2005) Molecular mapping of Fusarium head blight resistance in the winter wheat population Dream/Lynx. Theor Appl Genet 111:747–756
Schroeder HW, Christensen JJ (1963) Factors affecting resistance of wheat to scab caused by Giberella zeae. Phytopathology 53:831–838
Shen X, Ittu M, Ohm HW (2003) Quantitative trait loci conditioning resistance to Fusarium head blight in wheat line F201R. Crop Sci 43:850–857 Contribution from Purdue Univ., Agric. Res. Programs as Journal Article No. 16838
Snijders CHA (1990) Genetic variation for resistance to Fusarium head blight in bread wheat. Euphytica 50:171–179
Srinivasachary Gosman N, Steed A, Hollins TW, Bayles R, Jennings P, Nicholson P (2009) Semi-dwarfing Rht-B1 and Rht-D1 loci of wheat differ significantly in their influence on resistance to Fusarium head blight. Theor Appl Genet 118:695–702
Steed A, Chandler E, Thomsett M, Gosman N, Faure S, Nicholson P (2005) Identification of type I resistance to Fusarium head blight controlled by a major gene located on chromosome 4A of Triticum macha. Theor Appl Genet 111:521–529
Steiner B, Lemmens M, Griesser M, Scholz U, Schondelmaier J, Buerstmayr H (2004) Molecular mapping of resistance to Fusarium head blight in the spring wheat cultivar Frontana. Theor Appl Genet 109:215–224
Studer AJ, Doebley JF (2011) Do large effect QTL fractionate? A case study at the maize domestication QTL teosinte branched1. Genetics 188:673–681
Van Ooijen JW, Voorips RE (2001) Joinmap 3.0: Software for the calculation of genetic linkage maps. Plant Research International, Netherlands
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Waldron BL, Moreno-Sevilla B, Anderson JA, Stack RW, Frohberg RC (1999) RFLP mapping of QTL for Fusarium head blight resistance in wheat. Crop Sci 39:805–811
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang E, Maccaferri M, Salvi S, Milner S, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, IWGSC, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo M, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E (2014) Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421
Acknowledgments
This work was conducted as part of the ‘Integrated Strategy to Prevent Mycotoxin Risk’ (INSPYR) Project. This is a Sustainable Arable LINK project with funding from BBSRC, HGCA, DEFRA and RERAD (RD-207-3453). This work was also supported by the Biotechnology and Biological Sciences Research Council [BB/J004553/1 (BIO)] and by the EU FP7 MycoRed Project (GA 222690). The authors would like to thank Dr Peter Jack at RAGT Seeds Ltd. for population development and DNA extraction. We would also like to thank Djshwar Lateef for technical assistance with the field and polytunnel trials.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Buerstmayr.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2015_2542_MOESM1_ESM.bmp
Supplementary material 1 (BMP 1046 kb). Supplementary material 1 (BMP 1046 kb). Fig. S1: Histograms of predicted means for a AUDPC in JIC 2012, b %FHB in JIC 2012, c AUDPC in Polytunnel 2012, d %FHB in Polytunnel 2012, e AUPDC in JIC 2013, f %FHB in JIC 2013, AUDPC in CF 2013, and g %FHB in CF 2013
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Burt, C., Steed, A., Gosman, N. et al. Mapping a Type 1 FHB resistance on chromosome 4AS of Triticum macha and deployment in combination with two Type 2 resistances. Theor Appl Genet 128, 1725–1738 (2015). https://doi.org/10.1007/s00122-015-2542-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2542-9