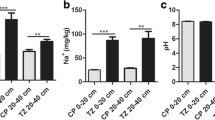
Abstract
Key message
QTL mapping in F 2 population [ V. luteola × V. marina subsp. oblonga ] revealed that the salt tolerance in V. marina subsp. oblonga is controlled by a single major QTL.
Abstract
The habitats of beach cowpea (Vigna marina) are sandy beaches in tropical and subtropical regions. As a species that grows closest to the sea, it has potential to be a gene source for breeding salt-tolerant crops. We reported here for the first time, quantitative trait loci (QTLs) mapping for salt tolerance in V. marina. A genetic linkage map was constructed from an F2 population of 120 plants derived from an interspecific cross between V. luteola and V. marina subsp. oblonga. The map comprised 150 SSR markers. The markers were clustered into 11 linkage groups spanning 777.6 cM in length with a mean distance between the adjacent markers of 5.59 cM. The F2:3 population was evaluated for salt tolerance under hydroponic conditions at the seedling and developmental stages. Segregation analysis indicated that salt tolerance in V. marina is controlled by a few genes. Multiple interval mapping consistently identified one major QTL which can explain about 50 % of phenotypic variance. The flanking markers may facilitate transfer of the salt tolerance allele from V. marina subsp. oblonga into related Vigna crops. The QTL for domestication-related traits from V. marina are also discussed.




Similar content being viewed by others
References
Arraouadi S, Badri M, Abdelly C, Huguet T, Aoudi ME (2012) QTL mapping of physiological traits associated with salt tolerance in Medicago truncatula recombinant inbred lines. Genomics 99:118–125
Ashraf MA, Foolad MR (2013) Crop breeding for salt tolerance in the era of molecular markers and marker-assisted selection. Plant Breed 132:10–20
Blair MW, Pedraza F, Buendia HF, Gaitán-Solís E, Beebe SE, Gepts P, Thome J (2003) Development of a genome-wide anchored microsatellite map of common bean (Phaseolus vulgaris L.). Theor Appl Genet 107:1362–1374
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
FAO (2005) Global network on integrated soil management for sustainable use of salt-affected soils. FAO land and plant nutrition management service, Rome
Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytol 179:945–963
Foolad MR (1999) Comparison of salt tolerance during seed germination and vegetative growth in tomato by QTL mapping. Genome 42:727–734
Foolad MR (2004) Recent advances in genetics of salt tolerance in tomato. Plant Cell Tissue Organ Cult 76:101–119
Gaitán-Solís E, Duque MC, Edwards KJ, Thome J (2002) Microsatellite repeats in common bean (Phaseolus vulgaris): isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop Sci 42:2128–2136
Greenway H, Munns H (1980) Mechanisms of salt tolerance in non halophytes. Annu Rev Plant Physiol 31:149–190
Han O, Kaga A, Isemura T, Wang XW, Tomooka N, Vaughan DA (2005) A genetic linkage map for azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi]. Theor Appl Genet 111:1288–1299
Isemura T, Kaga A, Konishi S, Ando T, Tomooka N, Han OK, Vaughan DA (2007) Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and comparison with other warm-season legumes. Ann Bot 100:1053–1071
Isemura T, Kaga A, Tomooka N, Shimizu T, Vaughan DA (2010) The genetics of domestication of rice bean, Vigna umbellata. Ann Bot 106:927–944
Isemura I, Kaga A, Tabata S, Somta P, Srinives P, Shimizu T, Jo U, Vaughan DA, Tomooka N (2012) Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS ONE 7(8):e41304
Kaga A, Vaughan DA, Tomooka N (2005) Molecular markers in Vigna improvement: understanding and using gene pools. In: Lorz H, Wanzel G (eds) Biotechnology in agriculture and forestry, vol 55., Molecular marker systemsBerlin, Springer, pp 171–187
Kaga A, Isemura T, Tomooka N, Vaughan DA (2008) The genetics of domestication of the azuki bean (Vigna angularis). Genetics 178:1013–1036
Kao CH, Zeng ZB, Teasdale RD (1999) Multiple interval mapping for quantitative trait loci. Genetics 152:1203–1216
Kearsey MJ, Pooni HS (1996) The genetical analysis of quantitative traits. Chapman and Hall, London
Kongjaimun A, Kaga A, Tomooka N, Somta P, Shimizu T, Shu Y, Vaughan DA, Srinives P (2012a) An SSR-based linkage map of yardlong bean [Vigna unguiculata ssp. sesquipedalis (L.) Verc.] and QTL analysis of pod length. Genome 55:81–92
Kongjaimun A, Kaga A, Tomooka N, Somta P, Vaughan DA, Srinives P (2012b) The genetics of domestication of yardlong bean [Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis]. Ann Bot 109:1185–1200
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lawn RJ, Cottrell A (1988) Wild mungbean and its relatives in Australia. Biologist 35:267–273
Lee SH (2012) Toward completion of genome sequence of mungbean. In: Abstract book, VI International Conference on Legume Genetics and Genomics, 2–7 October 2012, Hyderabad, India
Lee GJ, Boerma HR, Villagarcia MR, Zhou X, Carter TE Jr, Li Z, Gibbs MO (2004) A major QTL conditioning salt tolerance in S-100 soybean and descendant cultivars. Theor Appl Genet 109:1610–1619
Leonforte A, Sudheesh S, Cogan NOI, Salisbury PA, Nicolas ME, Materne M, Forster JW, Kaur S (2013) SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum sativum L.). BMC Plant Biol 13:161. doi:10.1186/1471-2229-13-161
Lewis G, Schrire B, Mackinder B, Lock M (2005) Legumes of the world. Kew, Royal Botanic Gardens
Li CD, Fatokun CA, Ubi B, Singh BB, Scoles GJ (2001) Determining genetic similarities and relationships among cowpea breeding lines and cultivars by microsatellite markers. Crop Sci 41:189–197
Lobato AKS, Santos Filho BG, Costa RCL, Goncalva-Vidigal MC, Moraes EC, Oliveira Neto CF, Rodriges VLF, Cruz FJR, Ferreira AS, Pita JD, Barreto AGT (2009) Morphological, physiological and biochemical responses during germination of the cowpea (Vigna unguiculata cv. Pitiuba) seeds under salt stress. World J Agric Sci 5:590–596
Lodhi MA, Ye GN, Weeden NF, Reisch BI (1994) A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol Biol Rep 12:6–13
Mano Y, Takeda K (1997) Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.). Euphytica 94:263–272
Maréchal R, Mascherpa JM, Stainer F (1978) Extude taxonomique d’un groupe d’espèces des genres Phaseolus et Vigna (V. unguiculata) sur la base de données morphologiques et polliniques, traitées pour l’analyse informatique. Boissiera 28:160–272
Mehra KL, Ibrahim AH (1989) Genetic resources in the Maldives. FAO/IBPGR Plant Genet Resour Newsl 75(76):42–43
Padulosi S, Ng NQ (1993) A useful and unexploited herb, Vigna marina (Leguminosae-Papilionideae) and the taxonomic revision of its genetic diversity. Bull Jard Bot Nat Belg 62:119–126
Palmer JL, Lawn RJ, Adkins SW (2002) An embryo-rescue protocol for Vigna interspecific hybrids. Aust J Bot 50:331–338
Quesada V, Martinez SG, Piqueras P, Ponce MR, Micol JL (2002) Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol 130:951–963
Rogers ME, Craig AD, Munns R et al (2005) The development of fodder plants for the salt-affected areas of southern and eastern Australia: an overview. Aust J Exp Agric 45:301–329
Samineni S (2010) Physiology, genetics and QTL mapping of salt tolerance in chickpea (Cicer arietinum L.). Ph.D. Thesis, The University of Western Australia
Sen NK, Bhowal JG (1960) Cytotaxonomic studies on Vigna. Cytologia 25:195–207
Smartt J (1978) The evaluation of pulse crops. Econ Bot 32:185–198
Somta P, Seehalak W, Srinives P (2009) Development, characterization and cross-species amplification of mungbean (Vigna radiata) genic microsatellite markers. Conserv Genet 10:1939–1943
Sonnante G, Spinosa A, Marangi A, Pignone D (1997) Isozyme and RAPD analysis of the genetic diversity within and between Vigna luteola and V. marina. Ann Bot 80:741–746
Tangphatsornruang S, Somta P, Uthaipaisanwong P, Chanprasert J, Sangsrakru D, Seehalak W, Sommanas W, Tragoonrung S, Srinives P (2009) Characterization of microsatellites and gene contents from genome shotgun sequences of mungbean (Vigna radiata (L.) Wilczek). BMC Plant Biol 9:137
Tomooka N, Vaughan DA, Moss H, Maxted N (2002) The Asian Vigna: genus Vigna subgenus Ceratotropis genetic resources. Kluwer Academic Publishers, Netherlands
Tomooka N, Kaga A, Isemura T, Vaughan D (2011) Vigna. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, Legume crops and forages. Springer, Berlin, pp 291–311
Tuyen DD, Lal SK, Xu DH (2010) Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theor Appl Genet 121:229–236
Van Ooijen JW (2006) JoinMap version 4.0, software for the calculation of genetic linkage maps. Kyazma B.V, Wageningen
Verdcourt B (1970) Studies in the Leguminosae-Papilionoideae for the ‘Flora of Tropical East Africa’, IV. Kew Bull 24:507–569
Verdcourt B (1971) Phaseoleae. In: Milne-Redhead E, Polhill RM (eds) Flora of tropical East Africa, Leguminosae (part 4), Papilionideae (2). Crown Agents for Overseas Governments, London, pp 625–627
Wang XW, Kaga A, Tomooka N, Vaughan DA (2004) The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi]. Theor Appl Genet 109:352–360
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. North Carolina State University, Raleigh
Wilson C, Liu X, Lesch SM, Suarez DL (2009) Growth response of major U.S. cowpea cultivars. I. Biomass accumulation and salt tolerance. Hortic Sci 41:225–230
Win KT, Oo AZ, Hirasawa T, Ookawa T, Yutaka H (2011) Genetic analysis of Myanmar Vigna species in response to salt stress at the seedling stage. Afr J Biotechnol 10:1615–1624
Yu K, Park J, Poysa V, Gepts P (2000) Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). J Hered 91:429–434
Zhang LP, Lin GY, Foolad MR (2003) QTL comparison of salt tolerance during seed germination and vegetative growth in a Lycopersicon esculentum × L. pimpinellifolium RIL population. Acta Hort 618:59–67
Zhang L, Wang S, Li H, Deng Q, Zheng A, Li S, Li P, Li Z, Wang J (2010) Effects of missing marker and segregation distortion on QTL mapping in F2 populations. Theor Appl Genet 121:1071–1082
Acknowledgments
This study was supported by JST, PRESTO to K. Naito, and by the Royal Golden Jubilee (RGJ) Ph.D. Program jointly funded by the Thailand Research Fund (TRF) and Kasetsart University to P. Srinives and S. Chankaew.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
All the experiments carried out in this study comply with the current laws of both Thailand and Japan.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by A. H. Schulman.
Electronic supplementary material
Below is the link to the electronic supplementary material.
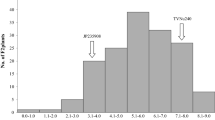
Supplementary Figure S1. Leave wilt scores using the scales of 1 to 9 for salt tolerance evaluation in the F2:3 population (V. luteola x V. marina subsp. oblonga) after being maintained at 300 mM for 30 days.
Rights and permissions
About this article
Cite this article
Chankaew, S., Isemura, T., Naito, K. et al. QTL mapping for salt tolerance and domestication-related traits in Vigna marina subsp. oblonga, a halophytic species. Theor Appl Genet 127, 691–702 (2014). https://doi.org/10.1007/s00122-013-2251-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2251-1