Abstract
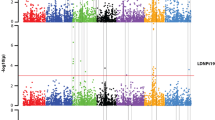
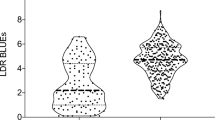
Fusarium head blight (FHB) and its associated mycotoxin, deoxynivalenol (DON), are the major biotic factors limiting cereal production in many parts of the world. A recent association mapping (AM) study of US six-row spring barley identified several modest effect quantitative trait loci (QTL) for DON and FHB. To date, few studies have attempted to verify the results of association analyses, particularly for complex traits such as DON and FHB resistance in barley. While AM methods use measures to control for the effects of population structure and multiple testing, false positive associations may still occur. A previous AM study used elite breeding germplasm to identify QTL for FHB and DON. To verify the results of that study, we evaluated the effects of the nine DON QTL using near-isogenic lines (NILs). We created families of contrasting homozygous haplotypes from lines in the original AM populations that were heterozygous for the DON QTL. Seventeen NIL families were evaluated for FHB and DON in three field experiments. Significant differences between contrasting NIL haplotypes were detected for three QTL across environments and/or genetic backgrounds, thereby confirming QTL from the original AM study. Several explanations for those QTL that were not confirmed are discussed, including the effect of genetic background and incomplete sampling of relevant haplotypes.


Similar content being viewed by others
References
Ahmad Naz A, Ehl A, Pillen K, Léon J (2012) Validation for root-related quantitative trait locus effects of wild origin in the cultivated background of barley (Hordeum vulgare L.). Plant Breed 3:392–398
Anderson JA, Stack RW, Liu S, Waldron BL, Fjeld AD, Coyne C, Moreno-Sevilla B, Mitchell Fetch J, Song QJ, Cregan PB, Frohlberg RC (2001) DNA markers for fusarium head blight resistance QTLs in two wheat populations. Theor Appl Genet 102:1164–1168
Austin DF, Lee M (1996) Genetic resolution and verification of quantitative trait loci for flowering and plant height with recombinant inbred lines of maize. Genome 39:957–968
Bai G, Shaner G (2004) Management and resistance in wheat and barley to Fusarium head blight. Ann Rev Phytopathol 42:135–161
Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48:1649–1664
Blake VC, Kling JG, Hayes PM, Jannink JL, Jillella SR, Lee J, Matthews DE, Shao S, Close TJ, Muehlbauer GJ, Smith KP, Wise RP, Dickerson JA (2012) The Hordeum toolbox: the barley CAP genotype and phenotype resource. Plant Genome 5:81–91
Brouwer DJ, St. Clair DA (2004) Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub-NILs. Theor Appl Genet 108:628–638
Buerstmayr H, Lemmens M, Hartl L, Doldi L, Steiner B, Stierschneider M, Ruckenbauer P (2002) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (Type II resistance). Theor Appl Genet 104:84–91
Buerstmayr H, Steiner B, Hartl L, Griesser M, Angerer N, Lengauer D, Miedaner T, Schneider B, Lemmens M (2003) Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor Appl Genet 107:503–508
Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128:1–26
Canci PC, Nduulu LM, Muehlbauer GJ, Dill-Macky R, Rasmusson DC, Smith KP (2004) Validation of quantitative trait loci for Fusarium head blight and kernel discoloration in barley. Mol Breed 14:91–104
Castro AJ, Chen X, Corey A, Filichkina T, Hayes PM, Mundt C, Richardson K, Sandoval-Islas S, Vivar H (2003) Pyramiding and validation of quantitative trait locus (QTL) alleles determining resistance to barley stripe rust. Crop Sci 43:2234–2239
Clear RM, Patrick SK, Nowicki T, Gaba D, Edney M, Babb JC (1997) The effect of hull removal and pearling on Fusarium species and trichothecenes in hulless barley. Can J Plant Sci 77:161–166
Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A, Stein N, Svensson JT, Wanamaker S, Bozdag S, Roose ML, Moscou MJ, Chao S, Varshney RK, Szucs P, Sato K, Hayes PM, Matthes DE, Kleinhofs A, Muehlbauer GJ, DeYoung J, Marshall DF, Madishetty K, Fenton RD, Condamine P, Graner A, Waugh R (2009) Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10:582
Cockram J, White J, Zuluaga DL, Smith D, Comadran J, Macaulay M, Luo Z, Kearsey MJ, Werner P, Harrap D, Tapsell C, Liu H, Hedley PE, Stein N, Schulte D, Steuernagel B, Marshall DF, Thomas WTB, Ramsay L, Mackay I, Balding DJ, Waugh R, O’Sullivan DM, AGOUEB Consortium (2010) Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc Natl Acad Sci USA 107:21611–21616
Dahleen LS, Agrama HA, Horsley RD, Steffenson BJ, Schwarz PB, Mesfin A, Franckowiak JD (2003) Identification of QTLs associated with Fusarium head blight resistance in Zhedar 2 barley. Theor Appl Genet 108:95–104
de la Pena RC, Smith KP, Capettini F, Muehlbauer GJ, Gallo-Meagher M, Dill-Macky R, Somers DA, Rassmuson DC (1999) Quantitative trait loci associated with resistance to Fusarium head blight and kernel discoloration in barley. Theor Appl Genet 99:561–569
Fasoula VA, Harris DK, Boerma HR (2004) Validation and designation of quantitative trait loci for seed protein, seed oil, and seed weight from two soybean populations. Crop Sci 44:1218–1225
Foolad MR, Zhang LP, Lin GY (2001) Identification and validation of QTLs for salt tolerance during vegetative growth in tomato by selective genotyping. Genome 44:444–454
Häberle J, Schmolke M, Schweizer G, Korzun V, Ebmeyer E, Zimmermann G, Hartl L (2007) Effects of two major Fusarium head blight resistance QTL verified in a winter wheat backcross population. Crop Sci 47:1823–1831
Hamblin MT, Jannink JL (2011) Factors affecting the power of haplotype markers in association studies. Plant Genome 4(2):145–153
Hamwieh A, Tuyen DD, Cong H, Benitez ER, Takahashi R, Xu DH (2011) Identification and validation of a major QTL for salt tolerance in soybean. Euphytica 179:451–459
Holland JB (2007) Genetic architecture of complex traits in plants. Curr Opin Plant Biol 10:156–161
Horsley RD, Schmierer D, Maier C, Kudrna D, Urrea CA, Steffenson BJ, Schwarz PB, Franckowiak JD, Green MJ, Zhang B, Kleinhofs A (2006) Identification of QTLs associated with Fusarium head blight resistance in barley accession CIho 4196. Crop Sci 46:145–156
Kaeppler SM, Phillips RL, Kim TS (1993) Use of near-isogenic lines derived by backcrossing or selfing to map qualitative traits. Theor Appl Genet 87:233–237
Kongprakhon P, Cuesta-Marcos A, Hayes PM, Richardson KL, Sirithunya P, Sato K, Steffenson B, Toojinda T (2009) Validation of rice blast resistance genes in barley using a QTL mapping population and near-isolines. Breeding Sci 59:341–349
Kraakman ATW, Martínez F, Mussiraliev B, Van Eeuwijk FA, Niks RE (2006) Linkage disequilibrium mapping of morphological, resistance, and other agronomically relevant traits in modern spring barley cultivars. Mol Breed 17:41–58
Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265:2037–2048
Landi P, Sanguineti MC, Salvi S, Giuliani S, Bellotti M, Maccaferri M, Conti S, Tuberosa R (2005) Validation and characterization of a major QTL affecting leaf ABA concentration in maize. Mol Breed 15:291–303
Legzdina L, Buerstmayr H (2004) Comparison of infection with Fusarium head blight and accumulation of mycotoxins in grain of hulless and covered barley. J Cereal Sci 40:61–67
Long AD, Langley CH (1999) The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res 9:720–731
Lorenz AJ, Hamblin MT, Jannink JL (2010) Performance of single nucleotide polymorphisms versus haplotypes for genome-wide association analysis in barley. PLoS ONE 5(11):e14079
Lorenz AJ, Smith KP, Jannink JL (2012) Potential and optimization of genomic selection for Fusarium head blight resistance in six-row barley. Crop Sci 52:1609–1621
Ma Z, Steffenson BJ, Prom LK, Lapitan NLV (2000) Mapping of quantitative trait loci for Fusarium head blight resistance in barley. Phytopathology 90:1079–1088
Massman J, Cooper B, Horsley R, Neate S, Dill-Macky R, Chao S, Dong Y, Schwarz P, Muehlbauer GJ, Smith KP (2011) Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol Breed 27:439–454
Mesfin A, Smith KP, Dill-Macky R, Evans CK, Waugh R, Gustus CD, Muehlbauer GJ (2003) Quantitative trait loci for Fusarium head blight resistance in barley detected in a two-rowed by six-rowed population. Crop Sci 43:307–318
Micic Z, Hahn V, Bauer E, Melchinger AE, Knapp SJ, Tang S, Schön CC (2005) Identification and validation of QTL for Sclerotinia midstalk rot resistance in sunflower by selective genotyping. Theor Appl Genet 111:233–242
Muehlbauer GJ, Specht JE, Thomas-Compton MA, Staswick PE, Bernard RL (1988) Near-isogenic lines-A potential resource in the integration of conventional and molecular marker linkage maps. Crop Sci 28:729–735
Muñoz-Amatriaín M, Castillo AM, Chen XW, Cistué L, Vallés MP (2008) Identification and validation of QTLs for green plant percentage in barley (Hordeum vulgare L.) anther culture. Mol Breed 22:119–129
Nduulu LM, Mesfin A, Muehlbauer GJ, Smith KP (2007) Analysis of the chromosome 2 (2H) region of barley associated with the correlated traits Fusarium head blight resistance and heading date. Theor Appl Genet 115:561–570
Pallotta MA, Warner P, Fox RL, Kuchel H, Jefferies SJ, Langridge P (2003) Marker assisted wheat breeding in the southern region of Australia. In: Pogna NE, Romano M, Pogna EA, Galerio G (eds) Proceedings of the tenth international wheat genetics symposium. Institutio Sperimentale per la Cerealcoltura, Rome, pp 789–791
Piepho HP (2005) Statistical tests for QTL and QTL-by-environment effects in segregating populations derived from line crosses. Theor Appl Genet 110:561–566
Prasad M, Kumar N, Kulwal PL, Röder MS, Balyan HS, Dhaliwal HS, Gupta PK (2003) QTL analysis for grain protein content using SSR markers and validation studies using NILs in bread wheat. Theor Appl Genet 106:659–667
Pumphrey MO, Bernardo R, Anderson JA (2007) Validating the Fhb1 QTL for Fusarium head blight resistance in near-isogenic wheat lines developed from breeding populations. Crop Sci 47:200–206
Romagosa I, Han F, Ullrich SE, Hayes PM, Wesenberg DM (1999) Verification of yield QTL through realized molecular marker-assisted selection responses in a barley cross. Mol Breed 5:143–152
Rostoks N, Ramsay L, MacKenzie K, Cardle L, Bhat PR, Roose ML, Svensson JT, Stein N, Varshney RK, Marshall DF, Graner A, Close TJ, Waugh R (2006) Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc Natl Acad Sci USA 103:18656–18661
Roy JK, Smith KP, Muehlbauer GJ, Chao S, Close TJ, Steffenson BJ (2010) Association mapping of spot blotch resistance in wild barley. Mol Breeding 26:243–256
SAS Institute (2008) SAS-statistical analysis software for windows, 9.2. SAS Institute, Cary, NC
Singh H, Prasad M, Varshney RK, Roy JK, Balyan HS, Dhaliwal HS, Gupta PK (2001) STMS markers for grain protein content and their validation using near-isogenic lines in bread wheat. Plant Breed 120:273–278
Smith KP, Evans CK, Dill-Macky R, Gustus C, Xie W, Dong Y (2004) Host genetic effect on deoxynivalenol accumulation in Fusarium head blight of barley. Phytopathology 94:766–771
Smith KP, Budde A, Dill-Macky R, Rasmusson DC, Schiefelbein E, Steffenson B, Wiersma JJ, Wiersma JV, Zhang B (2013) Registration of ‘Quest’ spring malting barley with improved resistance to Fusarium head blight. J Plant Regist 7:1–5. doi:10.3198/jpr2012.03.0200crc
Spaner D, Rossnagel BG, Legge WG, Scoles GJ, Eckstein PE, Penner GA, Tinker NA, Briggs KG, Falk DE, Afele JC, Hayes PM, Mather DE (1999) Verification of a quantitative trait locus affecting agronomic traits in two-row barley. Crop Sci 39:248–252
Steffenson BJ (2003) Fusarium head blight of barley: Impact, epidemics, management, and strategies for identifying and utilizing genetic resistance. In: Leonard KL, Bushnell WT (eds) Fusarium head blight of wheat and barley. The American Phytopathological Society, St. Paul, pp 241–295
Steffenson BJ, Smith KP (2006) Breeding barley for multiple disease resistance in the upper Midwest region of the USA. Czech J Genet Plant Breed 42:79–85
Stracke S, Haseneyer G, Veyrieras JB, Geiger HH, Sauer S, Graner A, Piepho HP (2009) Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theor Appl Genet 118:259–273
Tacke BK, Casper HH (1996) Determination of deoxynivalenol in wheat, barley, and malt by column cleanup and gas chromatography with electron capture detection. J AOAC Int 79:472–475
Tuinstra MR, Ejeta G, Goldsbrough PB (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95:1005–1011
Tuinstra MR, Ejeta G, Goldsbrough PB (1998) Evaluation of near-isogenic sorghum lines contrasting for QTL markers associated with drought tolerance. Crop Sci 38:835–842
Waldron BL, Moreno-Sevilla B, Anderson JA, Stack RW, Frohberg RC (1999) RFLP mapping of QTL for Fusarium head blight resistance in wheat. Crop Sci 39:805–811
Yun SJ, Gyenis L, Bossolini E, Hayes PM, Matus I, Smith KP, Steffenson BJ, Tuberosa R, Muelbauer GJ (2006) Validation of quantitative trait loci for multiple disease resistance in barley using advanced backcross lines developed with a wild barley. Crop Sci 46:1179–1186
Zhou W, Kolb FL, Bai G, Shaner G, Domier LL (2002) Genetic analysis of scab resistance QTL in wheat with microsatellite and AFLP markers. Genome 45:719–727
Zhu H, Gilchrist L, Hayes P, Kleinhofs A, Kudrna D, Liu Z, Prom L, Steffenson B, Toojinda T, Vivar H (1999) Does function follow form? Principal QTLs for Fusarium head blight (FHB) resistance are coincident with QTLs for inflorescence traits and plant height in a doubled-haploid population of barley. Theor Appl Genet 99:1221–1232
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Acknowledgments
We thank Ed Schiefelbein, Guillermo Velasquez, Karen Beaubien, Celeste Falcon, Leticia Kumar, Vikas Vikram, Ahmad Sallam, Richard Horsley, the University of Minnesota—Crookston field crews, and the University of Minnesota Small Grains Pathology lab for their contributions to phenotyping and field logistics. In addition, we thank Shiaoman Chao for SNP genotyping, Yanhong Dong for toxin analysis, and an anonymous reviewer for constructive suggestions on a prior version of this manuscript. Funding for this work was supported by grants from the National Institute of Food and Agriculture U.S. Dept. of Agriculture Agreement No. 2006-55606-16722 “Barley Coordinated Agricultural Project: Leveraging Genomics, Genetics, and Breeding for Gene Discovery and Barley Improvement”; the U.S. Wheat and Barley Scab Initiative U.S. Dept. of Agriculture ARS Agreement No. 59-0206-9-072, and the Rahr Foundation. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Department of Agriculture.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. J. Sillanpaa.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Navara, S., Smith, K.P. Using near-isogenic barley lines to validate deoxynivalenol (DON) QTL previously identified through association analysis. Theor Appl Genet 127, 633–645 (2014). https://doi.org/10.1007/s00122-013-2247-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2247-x