Abstract
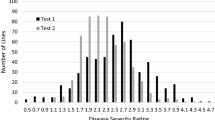
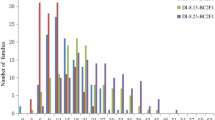
Diseases such as Fusarium wilt [Fusarium oxysporum f.sp. vasinfectum (FOV) Atk. Sny & Hans] represent expanding threats to cotton production. Integrating disease resistance into high-yielding, high-fiber quality cotton (Gossypium spp.) cultivars is one of the most important objectives in cotton breeding programs worldwide. In this study, we conducted a comprehensive analysis of gene action in cotton governing FOV race 4 resistance by combining conventional inheritance and quantitative trait loci (QTL) mapping with molecular markers. A set of diverse cotton populations was generated from crosses encompassing multiple genetic backgrounds. FOV race 4 resistance was investigated using seven parents and their derived populations: three intraspecific (G. hirsutum × G. hirsutum L. and G. barbadense × G. barbadense L.) F1 and F2; five interspecific (G. hirsutum × G. barbadense) F1 and F2; and one RIL. Parents and populations were evaluated for disease severity index (DSI) of leaves, and vascular stem and root staining (VRS) in four greenhouse and two field experiments. Initially, a single resistance gene (Fov4) model was observed in F2 populations based on inheritance of phenotypes. This single Fov4 gene had a major dominant gene action and conferred resistance to FOV race 4 in Pima-S6. The Fov4 gene appears to be located near a genome region on chromosome 14 marked with a QTL Fov4-C14 1 , which made the biggest contribution to the FOV race 4 resistance of the generated F2 progeny. Additional genetic and QTL analyses also identified a set of 11 SSR markers that indicated the involvement of more than one gene and gene interactions across six linkage groups/chromosomes (3, 6, 8, 14, 17, and 25) in the inheritance of FOV race 4 resistance. QTLs detected with minor effects in these populations explained 5–19 % of the DSI or VRS variation. Identified SSR markers for the resistance QTLs with major and minor effects will facilitate for the first time marker-assisted selection for the introgression of FOV race 4 resistance into elite cultivars during the breeding process.



Similar content being viewed by others
References
Armstrong JK, Armstrong GM (1958) A race of the cotton wilt Fusarium causing wilt of Yelredo soybean and flue-cured tobacco. Plant Dis Rep 42:147–151
Armstrong GM, Armstrong JK (1960) American, Egyptian, and Indian cotton-wilt Fusaria: Their pathogenicity and relationship to other wilt fusaria. U.S. Dep. Agric. Tech, Bull 219
Armstrong GM, Armstrong JK (1978) A new race (race 6) of the cotton-wilt Fusarium from Brazil. Plant Dis Rep 62:421–423
Becerra LA, Lopez-Lavalle VJ, Gillespie VJ, Tate WA, Ellis MH, Stiller WN, Llewellyn DL, Wilson IW (2012) Molecular mapping of new sources of Fusarium wilt resistance in tetraploid cotton (Gossypium hirsutum L.). Mol Breed 30(2):1181–1191. doi:10.1007/s11032-012-9705-z
Bell AA (1984) Cotton protection practices in the USA and world. Section B: diseases. In: Kohel RJ, Lewis CF (ed) Cotton, agronomy monograph 24. ASA, CSSA, and SSSA, Madison, pp 288–309
Bolek Y, Bell AA, El-Zik KM, Thaxton PM, Magill CW (2005) Reaction of cotton cultivars and an F2 population to stem inoculation with isolates of Verticillium dahliae. J Phytopathol 153:269–273
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative traits mapping. Genetics 138:963–971
Davis RM, Colyer PD, Rothrock CS, Kochman JK (2006) Fusarium wilt of cotton: population diversity and implications for management. Plan Dis 90:692–703
DeVay JE (1986) Half a century dynamics and control of cotton diseases: Fusarium and Verticillium wilts. In: Proceedings of the beltwide cotton conference. National Cotton Council of America, Memphis, pp 35–41
Dighe ND, Robinson AF, Bell AA, Menz MA, Cantrell RG, Stelly DM (2009) Linkage mapping of resistance to reniform nematode in cotton following introgression from Gossypium longicalyx (Hutch. & Lee). Crop Sci 49:1151–1164
Fernandez D, Assigbetse K, Dubois MP, Geiger JP (1994) Molecular characterization of races and vegetative compatibility groups in Fusarium oxysporum f. sp. vasinfectum. Appl Environ Microbiol 60:4039–4046
Frelichowski JE, Palmer MB, Main D, Tomkins JP, Cantrell RG, Stelly DM, Yu J, Kohel RJ, Ulloa M (2006) Cotton genome mapping with new microsatellites from Acala ‘Maxxa’ BAC-ends. Mol Gen Genomics 275:479–491
Geiser DM, Jimenez-Gasco MM, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O’Donnel K (2004) FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur J Plant Pathol 110:473–479
Holmes EA, Bennett RS, Spurgeon DW, Colyer PD, Davis RM (2009) New genotypes of Fusarium oxysporum f. sp. vasinfectum from the southeastern United States. Plant Dis 93:1298–1304
Hutmacher B, Davis MR, Ulloa M, Wright S, Munk DS, Vargas RN, Roberts BA, Marsh BH, Keeley MP, Kim Y, Percy RG (2005) Fusarium in Acala and Pima cotton: symptoms and disease development. In: Proceedings of the beltwide cotton conference. National Cotton Council of America, Memphis, pp 245–246
Hutmacher B, Ulloa M, Wright SD, Davis MR, Keeley MP, Delgado R, Banuelos G, Marsh BH, Munk DS (2011) Fusarium Race 4: management recommendations for growers. In: Proceedings of the beltwide cotton conference. National Cotton Council of America, Memphis, pp 188–192
Ibrahim FM (1966) A new race of the cotton wilt Fusarium in the Sudan Gezira. Emp Cotton Grow Rev 43:296–299
Kim Y, Hutmacher RB, Davis RM (2005) Characterization of California isolates of Fusarium oxysporum f. sp. vasinfectum. Plant Dis 89:366–372
Kochman J, Swan L, Moore N, Bentley S, O’Neill W, Mitchell A, Obst N, Lehane J, Gulino L, Salmond G (2002) The Fusarium threat-are we making the progress? In: Proceeding of 11th cotton conference, August 13–15, 2002, Brisbane, pp 643–652
Kohel RJ, Yu J, Park YH, Lazo GR (2001) Molecular mapping and characterization of traits controlling fiber quality in cotton. Euphytica 121:163–172
McPherson RG, Jenkins JN, Watson C, McCarty JC (2004) Inheritance of root-knot nematode resistance in M-315 RNR and M78-RNR cotton. J Cotton Sci 8:154–161
Mohamed HA (1963) Inheritance of Resistance to Fusarium Wilt in some Egyptian Cottons. Empire Cotton Grow Rev 40:292–295
Ott L (1988) An introduction to statistical methods and data analysis. PWS-Kent Publ Comp, Boston, MA
Park YH, Alabady MS, Sickler B, Wilkins TA, Yu J, Stelly DM, Kohel RJ, El-Shihy OM, Cantrell RG, Ulloa M (2005) Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line (RIL) cotton population. Mol Gen Genomics 274:428–441
Rebouillat J, Dievart A, Vendeil JL, Escoute J, Giese G, Breitler JC, Gantet P, Espeout S, Guiderdoni E, Perin C (2009) Molecular genetics of rice root development. Rice 2:15–34
Roberts PA, Ulloa M (2010) Introgression of root-knot nematode resistance into tetraploid cottons. Crop Sci 50:940–951
Shepherd RL (1974) Transgressive segregation for root-knot nematode resistance in cotton. Crop Sci 14:872–875
Skovgaard K, Nirenberg HI, O’Donnell K, Rosendahl S (2001) Evolution of Fusarium oxysporum f. sp vasinfectum races inferred from multigene genealogies. Phytopathology 91:1231–1237
Smith AL, Dick JB (1960) Inheritance of resistance to Fusarium wilt in Upland and Sea Island cotton as complicated by nematodes under field conditions. Phytopathology 50:44–48
Smith SN, Ebbels DL, Garger RH, Kappelman AJ (1981) Fusarium wilt of cotton. In: Nelson PE, Toussoun TA, Cook RJ (eds) Fusarium: diseases, biology, and taxonomy. Pennsylvania State University, University Park, pp 29–38
Ulloa M, Hutmacher RB, Davis RM, Wright SD, Percy R, Marsh B (2006) Breeding for Fusarium wilt race 4 resistance in cotton under field and greenhouse conditions. J Cotton Sci 10:114–127
Ulloa M, Brubaker C, Chee P (2007) Cotton. In: Kole C (ed) Genome mapping and molecular breeding. Technical crops, vol 6. Springer, Berlin, pp 1–49
Ulloa M, Saha S, Yu JZ, Jenkins JN, Meredith WR Jr, Kohel RJ (2008) Lessons learned and challenges ahead of the cotton genome mapping. In: Proc. world cotton res conf, 1798. Lubbock. http://www.icac.org/meetings/wcrc/wcrc4/presentations/start.htm
Ulloa M, Percy R, Zhang J, Hutmacher RB, Wright SD, Davis RM (2009) Registration of four Pima cotton germplasm lines having good levels of Fusarium wilt Race 4 resistance with moderate yields and good fibers. J Plant Registr 3:198–202
Ulloa M, Wang C, Roberts PA (2010a) Gene action analysis by inheritance and quantitative trait loci mapping of resistance to root-knot nematodes in cotton. Plant Breed 129:541–550
Ulloa M, Hutmacher RB, Wright SD, Campbell B, Wallace T, Myers G, Bourland F, Weaver D, Chee P, Lubbers E, Thaxton P, Zhang J, Wayne S, Jones D (2010b) Beltwides’ elite Upland germplasm pool assessment of Fusarium wilt (FOV) races 1 & 4 in California. In: Proceedings of beltwide cotton conference. National Cotton Council of America, Memphis, pp 765–766
Ulloa M, Wang C, Hutmacher RB, Wright SD, Davis RM, Saski CA, Roberts PA (2011) Mapping Fusarium wilt race 1 genes in cotton by inheritance. QTL and sequencing composition. Mol Genet Genomics. doi:10.1007/s00438-011-0616-1
Van Ooijen JW (2004) MapQTL® 5. Software for the mapping of quantitative trait loci in experimental populations. Kyazma B.V, Wageningen
Van Ooijen JW (2006) JoinMap® 4.0 Software for the calculations of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen
Veech JA (1984) Cotton protection practices in the USA and World. In: Kohel RJ, Lewis CF (eds) Cotton. Agronomy monograph 24. ASA, CSSA, and SSSA. Madison, pp 309–329
Wang C, Roberts PA (2006) A Fusarium wilt resistance gene in Gossypium barbadense and its effect on root-knot nematode-wilt disease complex. Phytopathology 96:727–734
Wang C, Ulloa M, Roberts PA (2006) Identification and mapping of microsatellite markers linked to a root-knot nematode resistance gene (rkn1) in Acala NemX cotton (Gossypium hirsutum L.). Theor Appl Genet 112:770–777
Wang C, Ulloa M, Roberts PA (2008) A transgressive segregation factor (RKN2) in Gossypium barbadense for nematode resistance clusters with gene rkn1 in G. hirsutum. Mol Gen Genomics 279:41–52
Wang P, Su L, Qin L, Hu B, Guo W, Zhang T (2009) Identification and molecular mapping of Fusarium wilt resistant gene in upland cotton. Theor Appl Genet 119:733–739
Wang C, Ulloa M, Mullens TR, Yu JZ, Roberts PA (2012) QTL analysis for transgressive resistance to root-knot nematode in interspecific cotton (Gossypium spp.) progeny derived from susceptible parents. PLoS ONE 7(4):e34874
Weir SB (1996) Genetic data analysis II. Sinauer Associates, Inc., Sunderland
Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78:139–186
Yu Y, Yuan D, Liang S, Li X, Wang X, Lin Z, Zhang X (2011) Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. BMC Genomics 21:15
Yu J, Kohel RJ, Fang DD, Cho J, Van Deynze A, Ulloa M, Hoffman SM, Pepper AE, Stelly DM, Jenkins JN, Saha S, Kumpatla SP, Shah MR, Hugie WV, Percy RG (2012) A high-density simple sequence repeat and single nucleotide polymorphism genetic map of the tetraploid cotton genome. Genes Genomes Genet 2:43–58
Acknowledgments
This study was funded in part by a Reimbursable Agreement with Cotton Incorporated (CA State Support Committee), Cary, NC (ARIS Log Nos. 5303-05-00 0029833), a Cooperative Research Agreement with Cotton Incorporated, and a University of California Discovery Grant. The authors thank Drs. R. Kohel and J. Yu for providing the original seed source of the RIL population. The authors also thank S. Ellberg, M. Biggs, Z. Garcia, M. Keeley, R. Delgado, G. Banuelos, and many students for their help in evaluations. Use of greenhouse facilities of the University of California Kearney Research and Extension Center (Parlier, CA) and assistance of Fred Swanson and Laura Van der Staay is gratefully acknowledged. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture or University of California. The US Department of Agriculture and University of California are equal opportunity providers and employers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Xue.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ulloa, M., Hutmacher, R.B., Roberts, P.A. et al. Inheritance and QTL mapping of Fusarium wilt race 4 resistance in cotton. Theor Appl Genet 126, 1405–1418 (2013). https://doi.org/10.1007/s00122-013-2061-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2061-5