Abstract
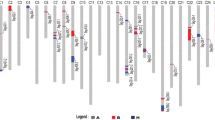
Gossypium hirsutum is a high yield cotton species that exhibits only moderate performance in fiber qualities. A promising but challenging approach to improving its phenotypes is interspecific introgression, the transfer of valuable traits or genes from the germplasm of another species such as G. barbadense, an important cultivated extra long staple cotton species. One set of chromosome segment introgression lines (CSILs) was developed, where TM-1, the genetic standard in G. hirsutum, was used as the recipient parent and the long staple cotton G. barbadense Hai7124 was used as the donor parent by molecular marker-assisted selection (MAS) in BC5S1–4 and BC4S1–3 generations. After four rounds of MAS, the CSIL population was comprised of 174 lines containing 298 introgressed segments, of which 86 (49.4%) lines had single introgressed segments. The total introgressed segment length covered 2,948.7 cM with an average length of 16.7 cM and represented 83.3% of tetraploid cotton genome. The CSILs were highly varied in major fiber qualities. By integrated analysis of data collected in four environments, a total of 43 additive quantitative trait loci (QTL) and six epistatic QTL associated with fiber qualities were detected by QTL IciMapping 3.0 and multi-QTL joint analysis. Six stable QTL were detected in various environments. The CSILs developed and the analyses presented here will enhance the understanding of the genetics of fiber qualities in long staple G. barbadense and facilitate further molecular breeding to improve fiber quality in Upland cotton.




Similar content being viewed by others
References
Chee P, Draye X, Jiang CX, Decanini L, Delmonte T, Brecdhauer R, Smith W, Paterson AH (2005a) Molecular dissection of interspecific variation between Gossypium hirsutum and G. barbadense (cotton) by a backcross-self approach: I. Fiber elongation. Theor Appl Genet 111:757–763
Chee P, Draye X, Jiang CX, Decanini L, Delmonte T, Brecdhauer R, Smith W, Paterson AH (2005b) Molecular dissection of phenotypic variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: III. Fiber length. Theor Appl Genet 111:772–781
Davasi A, Soller M (1995) Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 141:1199–1207
Dou BD, Hou BW, Wang F, Yang JB, Ni ZF, Sun QX, Zhang YM (2010) Further mapping of quantitative trait loci for female sterility in wheat (Triticum aestivum L.). Genet Res 92:63–70
Draye X, Chee P, Jiang CX, Decanini L, Delmonte T, Brecdhauer R, Smith W, Paterson AH (2005) Molecular dissection of interspecific variation between Gossypium hirsutum and G. barbadense (cotton) by a backcross-self approach: II. Fiber fineness. Theor Appl Genet 111:764–771
Ebitani T, Takeuchi Y, Nonoue Y, Yamamoto T, Takeuchi K, Yano M (2005) Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar ‘Kasalath’ in a genetic background of japonica elite cultivar ‘Koshihikari’. Breed Sci 55:65–73
Eshed Y, Zamir D (1994) A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79:175–179
Flagel LE, Udall JA, Nettleton D, Wendel JF (2008) Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol 6:6–16
Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88
Fridman E, Pleban T, Zamir D (2000) A recombination hotspot delimits a wild species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97:4718–4723
Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305:1786–1789
Fryxell PA (1992) A revised taxonomic interpretation of Gossypium L. (Malvaceae). Rheedea 2:108–165
Fukuoka S, Ebana K, Yamamoto T, Yano M (2010) Integration of genomics into rice breeding. Rice 3:131–137
Guo WZ, Zhang TZ, Zhu XF, Pan JJ (2005) Modified backcrossing pyramiding breeding with molecular marker-assisted selection and its application in cotton. Acta Agron Sin 31:963–970
Guo WZ, Cai CP, Wang CB, Zhao L, Wang L, Zhang TZ (2008) A preliminary analysis of genome structure and composition in Gossypium hirsutum. BMC Genomics 9:314–331
Han ZG, Wang CB, Song XL, Guo WZ, Guo JY, Li CH, Chen XY, Zhang TZ (2006) Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet 112:430–439
Hospital F (2002) Marker-assited backcross breeding: a case study in genotype buiding theory. In: Kang MS (ed) Quantitative genetics, genomics and plant breeding. CABI Publishing, Wallingford, pp 135–141
Hovav R, Udall JA, Chaudhary B, Rapp R, Flagel LE, Wendel JF (2008) Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploidy plant. Proc Natl Acad Sci USA 105:6191–6195
Jiang CX, Wright RJ, El-Zik K, Paterson AH (1998) Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc Natl Acad Sci USA 95:4419–4424
Kohel RJ, Lewis CF, Richmond TR (1970) Texas marker-1: description of a genetic standard for Gossypium hirsutum L. Crop Sci 10:670–671
Lacape JM, Nguyen TB, Courtois B, Belot JL, Giband M, Gourlot JP, Gawryziak G, Roques S, Hau B (2005) QTL analysis of cotton fiber quality trait using multiple Gossypium hirsutum × Gossypium barbadense backcross generations. Crop Sci 45:123–140
Lacape JM, Llewellyn D, Jacobs J, Arioli T, Becker D et al (2010) Meta-analysis of cotton fiber quality QTL across diverse environments in a Gossypium hirsutum × G. barbadense RIL population. BMC Plant Biol 10:132
Lin YR, Schertz KF, Paterson AH (1995) Comparative analysis of QTL affecting plants height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics 141:391–411
Lin Z, He D, Zhang X, Nie Y, Guo X, Feng C, Stewart JM (2005) Linkage map construction and mapping QTL for cotton fibre quality using SRAP, SSR and RAPD. Plant Breed 124:180–187
Liu JL, Sun JZ, Liu SQ, Zhang GY, Wang XF, Ma ZY (2000) Genetic studies of Verticillium wilt resistance among different types of Sea Island cottons. Acta Agronomica Sinica 26:315–321 (In Chinese)
Liu GM, Li WT, Zeng RZ, Zhang GQ (2003) Development of single segment substitution lines (SSSLs) of subspecies in rice. Chinese J Rice Sci 17:201–204 (In Chinese)
Luo ZW, Wu CI, Kearsey MJ (2002) Precision and high-resolution mapping of quantitative trait loci by use of recurrent selection, backcross or intercross schemes. Genetics 161:915–929
Ma XX, Ding YZ, Zhou BL, Guo WZ, Lv YH, Zhang TZ (2008) QTL mapping in A-genome diploid Asiatic cotton and their congruence analysis with AD-genome tetraploid cotton in genus Gossypium. J. Genet Genomics 35:751–762
Mei M, Syed NH, Gao W, Thaxton PM, Smith CW, Stelly DM, Chen ZJ (2004) Genetic mapping and QTL analysis of fiber-related traits in cotton (Gossypium). Theor Appl Genet 108:280–291
Neeraja CN, Maghirang-Rodriguez R, Pamplona A, Heuer S, Collard BCY, Septiningsih EM, Vergara G, Sanchez D, Xu K, Ismail AM, Mackill DJ (2007) A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet 115:767–776
Pan JJ, Zhang TZ, Kuai BK, Guo XP, Wang M (1994) Studies on the inheritance of resistance to Verticillium dahliae in cotton. J Nanjing Agric Univ 17:8–18 (In Chinese)
Paterson AH, Deverna JW, Lanini B, Tanksley SD (1990) Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes in an interspecies cross of tomato. Genetics 124:735–742
Paterson AH, Brubakerand C, Wendel JF (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep 11:122–127
Paterson AH, Saranga Y, Menz M, Jiang CX, Wright RJ (2003) QTL analysis of genotype × environment interactions affecting cotton fiber quality. Theor Appl Genet 106:384–396
Reinisch MJ, Dong J, Brubaker CL, Stelly DM, Wendel JF, Paterson AH (1994) A detailed RFLP map of cotton, Gossypium hirsutum × Gossypium barbadense: chromosome organization and evolution in a disomic polyploidy genome. Genetics 138:829–847
Rong JK, Feltus FA, Waghmare VN, Pierce GJ, Chee PW et al (2007) Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics 176:2577–2588
Shen XL, Guo WZ, Lu QX, Zhu XF, Yuan YL, Zhang TZ (2007) Genetic mapping of quantitative trait loci for fiber quality and yield trait by RIL approach in Upland cotton. Euphytica 155:371–380
Tian F, Zhu ZF, Zhang BS, Tan LB, Fu YC, Wang XK, Sun CQ (2006) Fine mapping of a quantitative trait locus for grain number per panicle from wild rice (Oryza rufipogon Griff.). Theor Appl Genet 113:619–629
Ulloa M, Meredith JR (2000) Genetic linkage map and QTL analysis of agronomic and fiber quality traits in an interspecific population. J Cotton Sci 4:161–170
Wan JM (2006) Perspectives of molecular design breeding in crops. Acta Agron Sin 32:455–462 (In Chinese)
Wang J, Wan X, Crossa J, Crouch J, Weng J, Zhai H, Wan J (2006a) QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet Res 88:93–104
Wang K, Song XL, Han ZG, Guo WZ, Yu JZ, Sun J, Pan JJ, Kohel RJ, Zhang TZ (2006b) Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor Appl Genet 113:73–80
Wang J, Wan X, Li H, Pfeiffer W, Crouch J, Wan J (2007) Application of identified QTL-marker associations in rice quality improvement through a design-breeding approach. Theor Appl Genet 115:87–100
Wendel JF, Olson PD, Stewart JM (1989) Genetic diversity, introgression and independent domestication of old world cultivated cottons. Am J Bot 76:1795–1806
Wu RL, Zeng ZB (2001) Joint linkage and linkage disequilibrium mapping in natural populations. Genetics 157:899–909
Xi ZY, He FH, Zeng RZ, Zhang ZM, Ding XH, Li WT, Zhang GQ (2006) Development of a wide population of chromosome single-segment substitution lines in the background of an elite cultivar of rice (Oryza sativa L.). Genome 49:476–484
Xu Z, Kohel RJ, Song GL, Cho JM, Alabady M et al (2008) Gene-rich islands for fiber development in the cotton genome. Genomics 92:173–183
Xu Y, Li HN, Li GJ, Wang X, Cheng LG, Zhang YM (2011) Mapping quantitative trait loci for seed size traits in soybean (Glycine max L. Merr.). Theor Appl Genet 122:581–594
Yamamoto T, Kuboki Y, Lin SY, Sasaki T, Yano M (1998) Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single Mendelian factors. Theor Appl Genet 97:37–44
Yang C, Guo WZ, Li GY, Gao F, Lin SS, Zhang TZ (2008) QTL mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Sci 174:290–298
Young ND, Tanksley SD (1989) Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor Appl Genet 77:95–101
Zhang YM (2006) Advances on methods of mapping QTL in plant. Chn Sci Bull 51:2809–2818
Zhang YM, Xu S (2005) A penalized maximum likelihood method for estimating epistatic effects of QTL. Heredity 95:96–104
Zhang J, Wu YT, Guo WZ, Zhang TZ (2000) Fast screening of microsatellite markers in cotton with PAGE/silver staining. Cotton Sci Sin 12:267–269
Zhang J, Guo WZ, Zhang TZ (2002) Molecular linkage map of allotetraploid cotton (Gossypium hirsutum L. × Gossypium barbadense L.) with a haploid population. Theor Appl Genet 105:1166–1174
Zhang YM, Mao YC, Xie CQ, Smith H, Lno L, Xu SZ (2005) Mapping QTL using naturally occurring genetic variance among commercial inbred lines of maize (Zea mays L.). Genetics 169:2267–2275
Acknowledgments
This study is supported by grants from the National Natural Science Foundation of China (30730067), the Priority Academic Program Development of Jiangsu Higher Education Institutions and Jiangsu Postdoctor Natural Science Foundation (0902022C).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Wang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, P., Zhu, Y., Song, X. et al. Inheritance of long staple fiber quality traits of Gossypium barbadense in G. hirsutum background using CSILs. Theor Appl Genet 124, 1415–1428 (2012). https://doi.org/10.1007/s00122-012-1797-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1797-7