Abstract
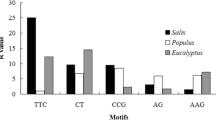
A collection of 5,659 expressed sequence tags (ESTs) from pineapple [Ananas comosus (L.) Merr.] was screened for simple sequence repeats (EST-SSRs) with motif lengths between 1 and 6 bp. Lower thresholds of 15, 7 and 5 repeat units were used to define microsatellites of the mono-, di-, and tri- to hexanucleotide repeat type, respectively. Based on these criteria, 696 SSRs were identified among 3,389 EST unigenes, together representing 2,840 kb. This corresponds to an average density of one SSR every 4.1 kb of non-redundant EST sequences. Dinucleotide repeats were most abundant (38.4% of all SSRs) followed by trinucleotide repeats (38.1%). Flanking primer pairs were designed for 537 EST-SSR loci, and 49 of these were screened for their functionality in 12 accessions of A. comosus, 14 accessions of 5 additional Ananas species and 1 species of Pseudananas. Distinct PCR products of the expected size range were obtained with 36 primer pairs. Eighteen loci analyzed in more detail were all polymorphic in pineapple, and primer pairs flanking these loci also generated PCR products from a wide range of genera and species from six subfamilies of the Bromeliaceae. The potential to reveal polymorphism in a heterologous target species was demonstrated in Deuterocohnia brevifolia (subfamily Pitcairnioideae).


Similar content being viewed by others
References
Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, Singh L (2007) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet 114:359–372. doi:10.1007/s00122-006-0440-x
Andersen JR, Lübberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560. doi:10.1016/j.tplants.2003.09.010
Ayers NM, McClung AM, Larkin PD, Bligh HFJ, Jones CA, Park WD (1997) Microsatellites and a single nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germplasm. Theor Appl Genet 94:773–781. doi:10.1007/s001220050477
Barbará T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C (2007) Cross-species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol 16:3759–3767. doi:10.1111/j.1365-294X.2007.03439.x
Benzing DH (2000) Bromeliaceae: profile of an adaptive radiation. Cambridge University, Cambridge
Boneh L, Kuperus P, van Tienderen PH (2003) Microsatellites in the bromeliads Tillandsia fasciculata and Guzmania monostachya. Mol Ecol Notes 3:302–303. doi:10.1046/j.1471-8286.2003.00432.x
Botella JR, Smith M (2008) Genomics of pineapple, crowning the king of tropical fruits. In: Moore PH, Ming R (eds) Plant genetics and genomics vol. 1: genomics of tropical crop plants. Springer, New York, pp 441–450. doi:10.1007/978-0-387-71219-2_18
Bray JR, Curtis JT (1957) An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349
Carlier JD, Reis A, Duval MF, Coppens d`Eeckenbrugge G, Leitão JM (2004) Genetic maps of RAPD, AFLP and ISSR markers in Ananas bracteatus and A. comosus using the pseudo-testcross strategy. Plant Breed 123:186–192. doi:10.1046/j.1439-0523.2003.00924.x
Carlier JD, Horta Sousa N, Espírito Santo T, Coppens d`Eeckenbrugge G, Leitão JM (2011) A genetic map of pineapple (Ananas comosus (L.) Merr.) including SCAR, CAPS, SSR and EST-SSR markers. Mol Breed. doi 10.1007/s11032-010-9543-9
Chen C, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257. doi:10.1007/s00122-006-0226-1
Collins JL (1960) The pineapple. Interscience Publishers, New York
Coppens d′Eeckenbrugge G, Leal F (2003) Morphology, anatomy and taxonomy. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The pineapple: botany, production and uses. CAB International, Wallingford, pp 13–32
Decroocq V, Favé MG, Hagen L, Bordenave L, Decroocq S (2003) Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet 106:912–922. doi:10.1007/s00122-002-1158-z
Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2010) Geneious v5.0, available from http://www.geneious.com [accessed 3 December 2010]
Duval M-F, Noyer JL, Perrier X, Coppens d′Eckenbrugge G, Hamon P (2001) Molecular diversity in pineapple assessed by RFLP markers. Theor Appl Genet 102:83–90
Duval MF, Buso GS, Ferreira FR, Noyer JL, Coppens d′Eeckenbrugge G, Hamon P, Ferreira ME (2003) Relationships in Ananas and other related genera using chloroplast DNA restriction site variation. Genome 46:990–1004. doi:10.1139/G03-074
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132. doi:10.1038/sj.hdy.6801001
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.1: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fraser LG, Harvey CF, Crowhurst RN, De Silva HN (2004) EST-derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet 108:1010–1016. doi:10.1007/s00122-003-1517-4
Gupta S, Prasad M (2009) Development and characterization of genic SSR markers in Medicago truncatula and their transferability in leguminous and non-leguminous species. Genome 52:761–771. doi:10.1139/G09-051
Heesacker A, Kishore VK, Gao W, Tang S, Kolkman JM, Gingle A, Matvienko M, Kozik A, Michelmore RM, Lai Z, Rieseberg LH, Knapp SJ (2008) SSRs and INDELs mined from the sunflower EST database: abundance, polymorphisms, and cross-taxa utility. Theor Appl Genet 117:1021–1029. doi:10.1007/s00122-008-0841-0
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rize, sorghum and wheat. Plant Mol Biol 48:501–510. doi:10.1139/G04-055
Kinsuat MJ, Kumar SV (2007) Polymorphic microsatellite and cryptic simple repeat sequence markers in pineapples (Ananas comosus var. comosus). Mol Ecol Notes 7:1032–1035. doi:10.1111/j.1471-8286.2007.01764.x
Kofler R, Schlötterer C, Lelley T (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23:1683–1685. doi:10.1093/bioinformatics/btm157
Kumpatla SP, Mukhopadhyay S (2005) Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48:985–998. doi:10.1139/G05-060
Luther HE (2008) An alphabetical list of bromeliad binomials, 11th edn. Bromeliad Society International, Mary Selby Botanical Gardens, Sarasota
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80. doi:10.1101/gr.10.1.72
Min XJ, Butler G, Storms R, Tsang A (2005) OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res 33:W677–W680. doi:10.1093/nar/gki394
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nature Genet 30:194–200. doi:10.1038/ng822
Moyle RL, Crowe ML, Ripi-Koia J, Fairbairn DJ, Botella JR (2005) PineappleDB: an online pineapple bioinformatics resource. BMC Plant Biol 5:21. doi:10.1186/1471-2229-5-21
Paggi GM, Palma-Silva T, Bered F, Cidade FW, Sousa ACB, Souza AP, Wendt T, Lexer C (2008) Isolation and characterization of microsatellite loci in Pitcairnia albiflos (Bromeliaceae), an endemic bromeliad from the Atlantic Rainforest, and cross-amplification in other species. Mol Ecol Resour 8:980–982. doi:10.1111/j.1755-0998.2008.02126.x
Palma-Silva C, Cavallari MM, Barbará T, Lexer C, Gimenes MA, Bered F, Bodanese-Zanettini MH (2007) A set of polymorphic microsatellite loci for Vriesea gigantea and Alcantarea imperialis (Bromeliaceae) and cross-amplification in other bromeliad species. Mol Ecol Notes 7:654–657. doi:10.1111/j.1471-8286.2006.01665.x
Pashley CH, Ellis JR, McCauley DE, Burke JM (2006) EST databases as a source for molecular markers: lessons from Helianthus. J Hered 97:381–388. doi:10.1093/jhered/es1013
Peakall R, Gilmore S, Keys W, Morgante M, Rafalski A (1998) Cross-species amplification of soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: implications for the transferability of SSRs in plants. Mol Biol Evol 15:1275–1287
Pinto LR, Oliveira KM, Ulian EC, Garcia AAF, de Souza AP (2004) Survey in the sugarcane expressed sequence tag database (SUCEST) for simple sequence repeats. Genome 47:795–804. doi:10.1139/G04-055
Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S, de Kochko A, Hamon P (2006) SSR mining in coffee tree EST databases: potential use of EST-SSRs as markers for the Coffea genus. Mol Gen Genomics 276:436–449. doi:10.1007/s00438-006-0153-5
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222. doi:10.1016/1360-1385(96)86898-1
Rohlf FJ (2000) NTSYSpc: numerical taxonomy and multivariate analysis system, ver. 2.1. Applied Biostatistic Inc, Port Jefferson
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: Methods in molecular biology. Humana Press, Totowa, pp 365–386
Sambrook J, Russell DW (2001) Molecular cloning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sarthou C, Boisselier-Dubayle MC, Lambourdière J, Samadi S (2003) Polymorphic microsatellites for the study of fragmented populations of Pitcairnia geyskesii L.B. Smith (Bromeliaceae), a specific saxicolous species of inselbergs in French Guiana. Mol Ecol Notes 3:221–223. doi:10.1046/j.1471-8286.2003.00404.x
Scaglione D, Acuadro A, Portis E, Taylor CA, Lanteri S, Knapp SJ (2009) Ontology and diversity of transcript-associated microsatellites mined from a globe artichoke EST database. BMC Genomics 10:454. doi:10.1186/1471-2164-10-454
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726. doi:10.1007/s001220051344
Smith L, Downs RJ (1979) Bromelioideae (Bromeliaceae). Monograph 14, part 3, Flora Neotropica. New York Botanical Garden, New York
Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W (2003) How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol 12:1339–1348. doi:10.1046/j.1365-294X.2003.01825.x
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422. doi:10.1007/s00122-002-1031-0
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55. doi:10.1016/j.tibtech.2004.11.005
Weising K, Nybom H, Wolff K, Kahl G (2005) DNA fingerprinting in plants: principles, methods, and applications, 2nd edn. CRC Press, Taylor & Francis Group, Boca Raton
You FM, Huo N, Gu YQ, Luo M-C, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD (2008) BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics 9:253. doi:10.1186/1471-2105-9-253
Yu J-K, Dake TM, Singh S, Benscher D, Li W, Gill B, Sorrells ME (2004) Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 47:805–818. doi:10.1139/G04-057
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation; a review. Mol Ecol 11:1–16. doi:10.1046/j.0962-1083.2001.01418.x
Zhang LY, Bernard M, Leroy P, Feuillet C, Sourdille P (2005) High transferability of bread wheat EST-derived SSRs to other cereals. Theor Appl Genet 111:677–687. doi:10.1007/s00122-005-2041-5
Acknowledgments
The plant material used in the present study was kindly provided by the Botanical Gardens of Berlin, Frankfurt, Göttingen, Hannover, Heidelberg, Kassel, Witzenhausen (all Germany) and Vienna (Austria), Prof. Dr. Ana Maria Benko Iseppon (University of Pernambuco, Recife, Brazil), Dr. Rafael B. Louzada (University of Sao Paulo, Brazil), Dipl.-Biol. Daniele Silvestro and Prof. Dr. Georg Zizka (Senckenberg Research Institute and Biodiversity and Climate Research Centre [BiK-F], Frankfurt), Dipl.-Biol. Nicole Schütz and Dipl.-Biol. Natascha Wagner (University of Kassel). We thank the ZFF (Zentrale Forschungsförderung) of the University of Kassel for funding the project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Nybom.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wöhrmann, T., Weising, K. In silico mining for simple sequence repeat loci in a pineapple expressed sequence tag database and cross-species amplification of EST-SSR markers across Bromeliaceae. Theor Appl Genet 123, 635–647 (2011). https://doi.org/10.1007/s00122-011-1613-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1613-9