Abstract
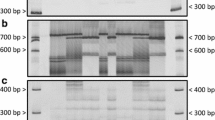
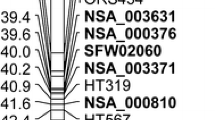
Rust is a serious fungal disease in the sunflower growing areas worldwide with increasing importance in North America in recent years. Several genes conferring resistance to rust have been identified in sunflower, but few of them have been genetically mapped and linked to molecular markers. The rust resistance gene R 4 in the germplasm line HA-R3 was derived from an Argentinean open-pollinated variety and is still one of most effective genes. The objectives of this study were to determine the chromosome location of the R 4 gene and the allelic relationship of R 4 with the R adv rust resistance gene. A total of 63 DNA markers previously mapped to linkage group (LG) 13 were used to screen for polymorphisms between two parental lines HA 89 and HA-R3. A genetic map of LG 13 was constructed with 21 markers, resulting in a total map length of 93.8 cM and an average distance of 4.5 cM between markers. Two markers, ZVG61 and ORS581, flanked the R 4 gene at 2.1 and 0.8 cM, respectively, and were located on the lower end of LG 13 within a large NBS-LRR cluster identified previously. The PCR pattern generated by primer pair ZVG61 was unique in the HA-R3 line, compared to lines HA-R1, HA-R4, and HA-R5, which carry other R 4 alleles. A SCAR marker linked to the rust resistance gene R adv mapped to LG 13 at 13.9 cM from the R 4 locus, indicating that R adv is not an allele of the R 4 locus. The markers tightly linked to the R 4 gene will facilitate gene pyramiding for rust resistance breeding of sunflower.



Similar content being viewed by others
References
Anderson PA, Lawrence GJ, Morrish BC, Ayliffe MA, Finnegan EJ, Ellis JG (1997) Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9:641–651
Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265:1856–1860
Burke JM, Tang S, Knapp SJ, Rieseberg LH (2002) Genetic analysis of sunflower domestication. Genetics 161:1257–1267
de Romano AB, Vázquez AN (2003) Origin of the Argentine sunflower varieties. Helia 26:127–136
Ellis JG, Lawrence G, Ayliffe M, Anderson P, Collins N et al (1997) Advances in the molecular genetic analysis of the flax–flax rust interaction. Annu Rev Phytopathol 35:271–291
Ellis JG, Dodds PN, Lawrence GJ (2007) Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol 45:289–306
Goulter KC (1990) Breeding of a rust differential sunflower line. In: Kochman JK (ed) Proceedings of the 8th Australian Sunflower Association workshop, 19–22 March 1990. Australian Sunflower Association, Toowoomba, pp 120–124
Gulya TJ (1985) Registration of five disease-resistant sunflower germplasms. Crop Sci 25:719–720
Gulya TJ, Markell S (2009) Sunflower rust status—2008. Race frequency across the midwest and resistance among commercial hybrids. http://www.sunflowernsa.com/uploads/Gulya_RustStatus_09.pdf
Gulya TJ, Masirevic S (1996) Inoculation and evaluation methods for sunflower rust. In: Proceedings of the 18th sunflower research workshop, Fargo, ND, 11–12 January 1996. National Sunflower Association, Bismarck, pp 31–38
Gulya TJ, Venette R, Venette JR, Lamey HA (1990) Sunflower rust. NDSU Ext. Ser. Bull. PP-998 Fargo, ND. http://www.ag.ndsu.edu/pubs/plantsci/rowcrops/pp998w.htm
Gulya TJ, Kong G, Brothers M (2000) Rust resistance in wild Helianthus annuus and variation by geographic origin. In: Proceedings of the 15th international sunflower conference, Toulouse, France, 12–15 June 2000, pp I38–42
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Ann Rev Plant Phys Plant Mol Biol 48:575–607
Horne EC, Kumpatla SE, Patterson KA, Gupta M, Thompson SA (2004) Improved high-throughput sunflower and cotton genomic DNA extraction and PCR fidelity. Plant Mol Biol Rep 22:83–84. Biology r 22:83–84
Huang L, Brooks SA, Fellers JP, Gill BS (2003) Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploidy genome of bread wheat. Genetics 164:655–664
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Ann Rev Phytopathol 39:285–312
Hulke BS, Miller JF, Gulya TJ (2010) Registration of the restorer oilseed sunflower germplasm RHA 464 possessing genes for resistance to downy mildew and sunflower rust. J Plant Regist 4:249–254
Islam MR, Mayo GME (1990) A compendium on host genes in flax conferring resistance to flax rust. Plant Breed 104:89–100
Islam MR, Shepherd KW (1991) Present status of genetics of rust resistance in flax. Euphytica 55:255–267
Jan CC, Gulya TJ (2006) Registration of a sunflower germplasm resistant to rust, downy mildew and virus. Crop Sci 46:1829
Jan CC, Quresh Z, Gulya TJ (2004) Registration of seven rust resistance sunflower germplasms. Crop Sci 44:1887–1888
Jones DA, Dickinson MJ, Balint-Kurti PJ, Dixon MS, Jones JDG (1993) Two complex resistance loci revealed in tomato by classical and RFLP mapping of the Cf-2, Cf-4, Cf-5, and Cf-9 genes for resistance to Cladosporium fulvum. Mol. Plant Microbe Interact 6:348–357
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic maps of experimental and natural population. Genomics 1:174–181
Lawson WR, Goulter KC, Henry RJ, Kong GA, Kochman JK (1998) Marker assisted selection for two rust resistance genes in sunflower. Mol Breed 4:227–234
Lawson WR, Jan CC, Shatte T, Smith L, Kong GA, Kochman JK (2010) DNA markers linked to the R 2 rust resistance gene in sunflower (Helianthus annuus L.) facilitate anticipatory breeding for this disease variant. Mol Breed. doi:10.1007/s11032-010-9506-1
Meyers BC, Chin DB, Shen KA, Sivaramakrishnan S, Lavelle DO et al (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10:1817–1832
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Miah MAJ, Sackston WE (1970) Genetics of host-pathogen interaction in sunflower. Phytoprotection 51:1–16
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Miller JF, Rodriguez RH, Gulya TJ (1988) Evaluation of genetic materials for inheritance of resistance to Race 4 rust in sunflower. In: Proceedings of the 12th international sunflower conference, Novi Sad, Yugoslavia, 25–29 July 1988. International Sunflower Association, Paris, pp 361-365
Prats E, Llamas MJ, Jorrin J, Rubiales D (2007) Constitutive coumarin accumulation on sunflower leaf surface prevents rust germ tube growth and appressorium differentiation. Crop Sci 47:1119–1124
Putt ED, Sackston WE (1963) Studies on sunflower rust. IV. Two genes, R 1 and R 2 for resistance in the host. Can J Plant Sci 43:490–496
Qi LL, Gulya TJ, Seiler GJ, Hulke BS, Vick BA (2011) Identification of resistance to new virulent races of rust in sunflowers and validation of DNA markers in the gene pool. Phytopathology 101:241–249
Quresh Z, Jan CC, Gulya TJ (1993) Resistance to sunflower rust and its inheritance in wild sunflower species. Plant Breed 110:297–306
Radwan O, Bouzidi MF, Vear F, Philippon J, Tourvieille de Labrouhe D, Nicolas P, Mouzeyar S (2003) Identification of non-TIR-NBS-LRR markers linked to the Pl5/Pl8 locus for resistance to downy mildew in sunflower. Theor Appl Genet 106:1438–1446
Radwan O, Gandhi S, Heesacker A, Whitaker B, Taylor C, Plocik A, Kesseli R, Kozik A, Michelmore R, Knapp SJ (2008) Genetic diversity and genomic distribution of homologs encoding NBS-LRR disease resistance proteins in sunflower. Mol Genet Genomics 280:111–125
Rashid KY (2006) Sunflower rust races in Manitoba. http://www.umanitoba.ca/afs/agronomists_conf/proceedings/2006/Rashid_sunflower_rust_races.pdf (verified 03 August 2010)
Richter TE, Ronald PC (2000) The evolution of disease resistance genes. Plant Mol Biol 42:195–204
Sackston WE (1962) Studies on sunflower rust. III. Occurrence, distribution, and significance of race Puccinia helianthi Schw. Can J Bot 40:1449–1458
Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, Kim HS et al (1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86:123–133
Sendall BC, Kong GA, Goulter KC, Aitken EAB, Thompson SM, Mitchell JHM, Kochman JK, Lawson W, Shatte T, Gulya TJ (2006) Diversity in the sunflower: Puccinia helianthi pathosystem in Australia. Australas Plant Path 35:657–670
Slabaugh MB, Yu JK, Tang SX, Heesacker A, Hu X, Lu GH, Bidney D, Han F, Knapp SJ (2003) Haplotyping and mapping a large cluster of downy mildew resistance gene candidates in sunflower using multilocus intron fragment length polymorphisms. Plant Biotechnol J 1:167–185
Song WY, Pi LY, Wang GL, Gardner J, Holsten T et al (1997) Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9:1279–1287
Stakman EC, Stewart DM, Loegering WQ (1962) Identification of physiological races of Puccinia graminis var. tritici. USDA-ARS Bulletin E-617
Tang S, Yu JK, Slabaugh MB, Shintani DK, Knapp SJ (2002) Simple sequence repeat map of the sunflower genome. Theor Appl Genet 105:1124–1136
Tang S, Kishore VK, Knapp SJ (2003) PCR-multiplexes for a genome-wide framework of simple sequence repeat marker loci in cultivated sunflower. Theor Appl Genet 107:6–19
Trevet IW, Rawson AJ, Cherry E, Saxon RB (1951) A method for the collection of microscopic particles. Phytopathology 41:282–285
Wei F, Wing RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14:1903–1917
Yahiaoui N, Arichumpa P, Dudler R, Keller B (2004) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37:528–538
Yang SM, Antonelli EF, Luciano A, Luciani ND (1986) Reactions of Argentine and Australian sunflower rust differentials to four North American cultures of Puccinia helianthi from North Dakota. Plant Dis 70:883–886
Yang SM, Dowler WM, Luciano A (1989) Gene Pu 6 : a new gene in sunflower for resistance to Puccinia helianthi. Phytopathology 79:474–477
Yang S, Jiang K, Araki H, Ding J, Yang YH, Tian D (2007) A molecular isolation mechanism associated with high intra-specific diversity in rice. Gene 394:87–95
Yu JK, Tang S, Slabaugh MB, Heesacker A, Cole G, Herring M et al (2003) Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci 43:367–387
Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D (2004) Genome-wide identification of NBS genes in rice reveals significant expansion of divergent non-TIR NBS Genes. Mol Genet Genomics 271:402–415
Acknowledgments
We thank Drs. Justin Faris and Chao-Chien Jan for critical review of the manuscript, and Angelia Hogness and Megan Ramsett for excellent technical assistance both in the lab and greenhouse. This project was supported by the USDA-ARS CRIS Project No. 5442-21000-034-00D.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Bervillé.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
Rights and permissions
About this article
Cite this article
Qi, L.L., Hulke, B.S., Vick, B.A. et al. Molecular mapping of the rust resistance gene R 4 to a large NBS-LRR cluster on linkage group 13 of sunflower. Theor Appl Genet 123, 351–358 (2011). https://doi.org/10.1007/s00122-011-1588-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1588-6