Abstract
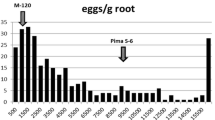
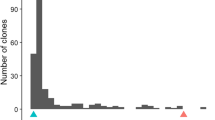
The identification and utilization of a high-level of host plant resistance is the most effective and economical approach to control root-knot nematode (Meloidogyne incognita). In an earlier study, we identified a major quantitative trait locus (QTL) for resistance to root-knot nematode in the M-120 RNR Upland cotton line (Gossypium hirsutum L.) of the Auburn 623 RNR source. The QTL is located in a 12.9-cM interval flanked by the two SSR markers CIR069 and CIR316 on the distal segment of chromosome 11. To construct a fine map around the target region, a bulked segregation analysis was performed using two DNA pools consisting of five individuals, with each being homozygous for the two parental alleles. From a survey of 1,152 AFLP primer combinations, 9 AFLP markers closely linked to the target region were identified. By screening an additional 1,221 F2 individuals developed from the initial mapping population, the Mi-C11 locus was delimited to a 3.6-cM interval flanked by the SSR marker CIR069 and the AFLP marker E14M27-375. These results further elucidate the genetic fine structure of the Mi-C11 locus and provide the basis for map-based isolation of the nematode resistance gene in M-120 RNR.



Similar content being viewed by others
References
Basten CJ, Weir BS, Zeng ZB (2001) QTL cartographer, Version 1.15. Department of Statistics, North Carolina State University, Raleigh
Bolek Y, El-Zik KM, Pepper AE, Bell AA, Magill CM, Thaxton PM, Reddy OUK (2005) Mapping of verticillum wilt resistance genes in cotton. Plant Sci 168:1581–1590
Bridge J, Page SLR (1980) Estimation of root-knot nematode infestation levels on roots using a rating chart. Trop Pest Manage 26:296–298
Chee PW, Rong J, Williams-Coplin D, Schulze SR, Paterson AH (2004) EST derived PCR-based markers for functional gene homologues in cotton. Genome 47:449–462
Cotton Disease Loss Estimate Committee (2008) Cotton disease loss estimate committee report. In: Proceedings of the Beltwide Cotton Conference. Nashville, TN, Jan 8–11. National Cotton Council of America, Memphis, TN, pp 102–103
Dighe ND, Robinson AD, Bell AA, Menz MA, Cantrell RG, Stelly DM (2009) Linkage mapping of resistance to reniform nematode in cotton following introgression from Gossypium longicalyx. Crop Sci 49:1151–1164
Fassuliotis G, Deakin JR, Hoffman (1970) Root-knot nematode resistance in snap beans: breeding and nature of resistance. J Am Soc Hortic Sci 95:640–645
Ferreira AM, Da Silva F, Silva L, Cruz CD (2006) Estimating the effects of population size and type on the accuracy of genetic maps. Genet Mol Biol 29:187–192
Harris DK, Boerma HR, Hussey RS, Finnerty SL (2003) Additional sources of soybean germplasm resistant to two species of root-knot nematode. Crop Sci 43:1848–1851
He Y, Iqbal N, Shen X, Davis RF, Chee P (2010) QTL mapping of resistance to root-knot nematode in the wild cotton germplasm line M-495RNR. In: Proceedings of the Beltwide Prod Res Conference, New Orleans, LA, Jan 9–12 2010. National Cotton Council of America, Memphis, TN
Hussey RS, Barker KR (1973) A comparison of methods of collecting inocula of Meloidogyne spp. including a new technique. Plant Dis Rep 57:1025–1028
Jenkins JN, Creech RG, Tang B, Lawrence GW, McCarty JC (1995) Cotton resistance to root-knot nematode: II. Post-penetration development. Crop Sci 35:369–373
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
McPherson MG, Jenkins JN, McCarty JC, Watson CE (1995) Combining ability analysis of root-knot nematode resistance in cotton. Crop Sci 35:373–375
McPherson MG, Jenkins JN, Watson CE, McCarty JC (2004) Inheritance of root-knot nematode resistance in M-315 RNR and M78-RNR cotton. J Cotton Sci 8:154–161
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis. A rapid method to detect markers in specific genomic regions by using segregating population. Proc Natl Acad Sci USA 88:6553–6558
Niu C, Hinchliffe DJ, Cantrell RG, Wang C, Phillip AR, Zhang J (2007) Identification of molecular markers associated with Root-knot nematode resistance in upland cotton. Crop Sci 47:951–960
Paterson AH, DeVern JW, Lanini B, Tanksley SD (1989) Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in a interspecific cross of tomato. Genetics 124:735–742
Roberts PA, Ulloa M (2010) Introgression of root-knot nematode resistance into tetraploid cotton. Crop Sci 50:940–951
Roberts PA, Matthews WC, Ehlers JD, Helms D (2008) Genetic determinants of differential resistance to root-knot nematode reproduction and galling in lima bean. Crop Sci 48:553–560
Robinson AF, Bowman DT, Cook CG, Jenkins JN, Jones JE, May OL, Oakley SR, Oliver MJ, Roberts PA, Robinson M, Smith CW, Starr JL, Stewart JM (2001) Nematode Resistance. In: Kirkpatrick TL, Rothrock CS (eds) Compendium of cotton diseases. The American Phytopathological Society, St. Paul, pp 68–72
Romano GB, Sacks EJ, Stetina SR, Robinson AG, Fang DD, Gutierrez OA, Scheffler JA (2009) Identification and genomic location of a reniform nematode (Rotylenchulus reniformis) resistance locus (Renari) introgressed from Gossypium aridum into upland cotton (G. hirsutum). Theor Appl Genet 120:139–150
Shen X, Van Becelaere G, Kumar P, Davis RF, May OL, Chee P (2006) QTL mapping for resistance to root-knot nematodes in the M-120 RNR Upland cotton line (Gossypium hirsutum L.) of the Auburn 623 RNR source. Theor Appl Genet 113:1539–1549
Shepherd RL (1974a) Registration of Auburn 623RNR cotton germplasm. Crop Sci 14:911
Shepherd RL (1974b) Transgressive segregation for root-knot nematode resistance in cotton. Crop Sci 14:872–875
Shepherd RL (1979) A quantitative technique for evaluating cotton for root-knot nematode resistance. Phytopathology 69:427–430
Shepherd RL, McCarty JC, Jenkins JN (1996) Registration of nine cotton germplasm lines resistance to root-knot nematode. Crop Sci 36:820
Smith AL (1941) The reaction of cotton varieties to Fusarium wilt and root-knot nematode. Phytopathology 31:1099–1107
Starr JL, Koenning SR, Eirkpatrick TL, Robinson AF, Roberts PA, Nichols RL (2007) The future of nematode management in cotton. J Nematol 39:283–294
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang C, Ulloa M, Roberts P (2006a) Identification and mapping of microsatellite markers linked to a root-knot nematode resistance gene (rkn1) in Acala NemX cotton. Theor Appl Genet 112:770–777
Wang K, Song X, Han Z, Guo W, Yu JZ, Sun J, Pan J, Kohel RJ, Zhang T (2006b) Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor Appl Genet 113:73–80
Wang CL, Ulloa M, Roberts PA (2008) A transgressive segregation factor (RKN2) in Gossypium barbadense for nematode resistance clusters with gene rkn1 in Gossypium hirsutum. Mol Genet Genomics 279:41–52
Ynturi P, Jenkins JN, McCarty JC, Gutirerez OA, Saha S (2006) Association of root-knot nematode resistance genes with simple sequence repeat markers on two chromosomes in cotton. Crop Sci 46:2670–2674
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhang J, Guo WZ, Zhang TZ (2002) Molecular linkage map of allotetraploid cotton (Gossypium hirsutum L. × Gossypium barbadense L.) with a haploid population. Theor Appl Genet 105:1166–1174
Acknowledgments
We thank the Zhang lab for SSR primer sequences; Jennifer McCurdy and Rippy Singh for technical assistance; Pawan Kumar for valuable comments; and gratefully acknowledge the financial support from the Georgia Cotton Commission and Cotton Incorporated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Schön.
Rights and permissions
About this article
Cite this article
Shen, X., He, Y., Lubbers, E.L. et al. Fine mapping QMi-C11 a major QTL controlling root-knot nematodes resistance in Upland cotton. Theor Appl Genet 121, 1623–1631 (2010). https://doi.org/10.1007/s00122-010-1415-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1415-5