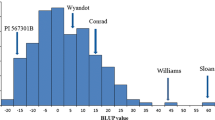
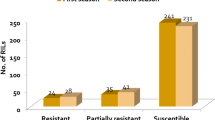
Abstract
Phytophthora root rot (PRR) of soybean (Glycine max (L.) Merr.) is the second most important cause of yield loss by disease in North America, surpassed only by soybean cyst nematode (Wrather et al. in Can J Plant Pathol 23:115–121, 2001). Tolerance can provide economically useful disease control, conditioning partial resistance of soybean to PRR. The aims of this study were to identify new quantitative trait loci (QTL) underlying tolerance to PRR, and to evaluate the effects of pyramided or stacked loci on the level of tolerance. A North American cultivar ‘Conrad’ (tolerant to PRR) was crossed with a northeastern China cultivar ‘Hefeng 25’ (tolerant to PRR). Through single-seed descent, 140 F2:5 and F2:6 recombinant inbred lines were advanced. A total of 164 simple sequence repeat (SSR) markers were used to construct a genetic linkage map. The percentage of seedling death was measured over 2 years (2007 and 2008) in the field at four naturally infested locations in Canada and China following additional soil infestation and in the greenhouse following inoculation with Phytophthora sojae isolate. A total of eight QTL underlying tolerance to PRR were identified, located in five linkage groups (F, D1b+w, A2, B1, and C2). The phenotypic variation contributed by the loci ranged from 4.24 to 27.98%. QPRR-1 (anchored in the interval of SSR markers Satt325 and Satt343 of LG F), QPRR-2 (anchored in the interval of Satt005 and Satt600 of LG D1b+w), and QPRR-3 (anchored in the interval of Satt579 and Sat_089 of LG D1b+w) derived their beneficial allele from ‘Conrad’. They were located at chromosomal locations known to underlie PRR tolerance in diverse germplasm. Five QTL that derived beneficial alleles from ‘Hefeng 25’ were identified. The QTL (QPRR-1 to QPRR-7) that were detected across at least three environments were selected for loci stacking and to analyze the relationship between number of tolerance loci and disease loss percentage. The accumulation of tolerance loci was positively correlated with decreases in disease loss percentage. The pyramid of loci underlying tolerance to PRR provided germplasm useful for crop improvement by marker-assisted selection and may provide durable cultivar tolerance against the PRR disease.

Similar content being viewed by others
References
Abney T, Melgar J, Richards T, Scott D, Grogan J, Young J (1997) New races of Phytophthora sojae with Rps1-d virulence. Plant Dis 81:653–655
Basten CJ, Weir BS, Zeng ZB (1996) QTL cartographer. North Carolina State University, NC
Beavis WD (1998) QTL analyses: power, precision, and accuracy. In: Paterson AH (ed) Molecular dissection of complex traits. CRC Press, Boca Raton, pp 145–162
Bernard RL, Smith PE, Kaufmann MJ, Schmitthenner AF (1957) Inheritance of resistance to phytophthora root rot and stem rot in the soybean. Agron J 49:391
Bhat R, Schmitthenner A (1993) Genetic crosses between physiologic races of Phytophthora sojae. Exp Mycol 17:122–129
Blum A, Klueva N, Nguven HT (2001) Wheat cellular thermo tolerance is related to yield under heat stress. Euphytica 117:117–123
Burnham K, Dorrance A, Francis D, Fioritto R, St. Martin S (2003a) Rps8, a new locus in soybean for resistance to Phytophthora sojae. Crop Sci 43:101–105
Burnham K, Dorrance A, VanToai T, St. Martin S (2003b) Quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Crop Sci 43:1609–1617
Churchill RW, Doerge GA (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cregan P, Jarvik T, Bush A, Shoemaker R, Lara K, Kahler A, Kaya N, VanToai T, Lohnes D, Chung J, Specht J (1999) An integrated genetic linkage map of the soybean genome. Crop Sci 39:1464–1490
Dorrance AE, McClure SA, St. Martin SK (2003) Effect of partial resistance on Phytophthora stem rot incidence and yield of soybean in Ohio. Plant Dis 87:308–312
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Drenth A, Whisson S, MacLean D, Irwin J, Obst N, Riley M (1996) The evolution of Phytophthora sojae in Australia. Phytopathology 86(2):163–169
Fehr WR, Cianzio SR, Voss BK, Schultz SP (1989) Registration of ‘Conrad’ soybean. Crop Sci 29:830
Han YP, Teng WL, Yu KF, Poysa V, Anderson T, Qiu LJ, Lightfoot DA, Li WB (2008) Mapping QTL tolerance to Phytophthora root rot in soybean using microsatellite and RAPD/SCAR derived markers. Euphytica 162:231–239
Hartman G, Sinclair J, Rupe J (1999) A compendium of soybean diseases, 4th edn. The American Phytopathological Society, St Paul, p 100
Hildebrand A (1959) A root and stalk rot of soybean caused by Phytophthora megaspema var. sojae var. Can J Bot 37:927–957
Irwin J, Cahill D, Drenth A (1995) Phytophthora in Australia. Aust J Agric Res 46:1311–1337
Jee H, Kim W, Cho W (1998) Occurrence of Phytophthora root rot on soybean (Glycine max) and identification of the causal fungus. Crop Prot 40:16–22
Kaufmann MJ, Gerdemann W (1958) Root and stem rot of soybean caused by Phytophthora sojae n. sp. Phytopathology 48:201–208
Lander ES, Green P, Abrahamson J, Baarlow A, Daly MJ, Lincoln SE, Newburg L (1987) MapMaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Leitz R, Hartman G, Pederson W, Nickell C (2000) Races of Phytophthora sojae on soybean in Illinois. Plant Dis 84:487
Malvick D, Grunden E (2004) Traits of soybean-infecting Phytophthora populations from Illinois agricultural Welds. Plant Dis 88:1139–1145
Mundt CC (2000) Use of multiline cultivars and cultivar mixtures for disease management. Annu Rev Phytopathol 40:381–410
Njiti V, Gray L, Lightfoot DA (1997) Rate-reducing resistance to Fusarium solani f.sp. phaseoli [nee: glycines] underlies field resistance to soybean sudden-death syndrome (SDS). Crop Sci 37:1–12
Njiti V, Johnson JE, Torto TA, Gray LE, Lightfoot DA (2001) Inoculum rate influences selection for field resistance to soybean sudden death syndrome in the greenhouse. Crop Sci 41:1726–1731
Njiti V, Meksem K, Iqbal MJ, Johnson JE, Kassem MA, Zobrist KF, Kilo VY, Lightfoot DA (2002) Common loci underlie field resistance to soybean sudden death syndrome in Forrest, Pyramid, Essex, and Douglas. Theor Appl Genet 104:294–300
Prabhu RR, Njiti VN, Johnson JE, Schmidt ME, Klein RJ, Lightfoot DA (1999) Selecting soybean cultivars for dual resistance to cyst nematode sudden death syndrome with two DNA markers. Crop Sci 39:982–987
Ryley M, Obst N, Irwin J, Drenth A (1998) Changes in the racial composition of Phytophthora sojae in Australia between 1979 and 1996. Plant Dis 82:1048–1054
Schmitthenner AF (1989) Phytophthora root rot. In: Sinclair JB, Backman PA (eds) Compendium of soybean diseases. APS Press, St Paul, pp 35–38
Song Q, Marek L, Shoemaker R, Lark K, Concibido V, Delannay X, Specht J, Cregan P (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128
Su Y, Yao S (1993) The discovery and biological characteristics studies of Phytophthora megasperma f.sp.glycinea on soybean in China. Acta Phytopathol Sin 23:341–347
Sugimoto T, Aino M, Sugimoto M, Watanabe K (2005) Reduction of Phytophthora stem rot disease on soybeans by the application of CaCl2 and Ca(NO3)2. J Phytopathol 153:536–543
Tooley PW, Grau CR (1982) Identification and quantitative characterization of rate-reducing resistance to Phytophthora megasperma f. sp. glycinea in soybean seedlings. Phytopathology 72:727–733
Tooley PW, Grau CR (1984a) Field characterization of rate-reducing resistance to Phytophthora megasperma f. sp. glycinea in soybean. Phytopathol 74:1201–1208
Tooley PW, Grau CR (1984b) The relationship between rate-reducing resistance to Phytophthora megasperma f.sp. glycinea and yield of soybean. Phytopathol 74:1209–1216
Trigiano RN, Caetano-Anolles G (1998) Laboratory exercises on DNA amplification fingerprinting for evaluating the molecular diversity of horticultural species. HortTechnology 8:413–423
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTL. Heredity 93:77–78
Weng CR, Yu KF, Anderson T, Poysa V (2007) A quantitative trait locus influencing tolerance to Phytophthora root rot in the soybean cultivar ‘Conrad’. Euphytica 158:81–86
Wrather JA, Anderson TR, Arsyad DM, Tan Y, Ploper LD, Porta-Puglia A, Ram HH, Yorinori JT (2001) Soybean disease loss estimates for the top ten soybean producing countries in 1998. Can J Plant Pathol 23:115–121
Yuan J, Njiti VN, Meksem K, Iqbal MJ, Triwitayakorn K, Kassem MA, Davis GT, Schmidt ME, Lightfoot DA (2002) Quantitative trait loci in two soybean recombinant inbred line populations segregating for yield and disease resistance. Crop Sci 42:271–277
Zeng Z (1993) Theoretical basis of separation of multiple linked gene effects on mapping quantitative trait loci. Proc Natl Acad Sci USA 90:10972–10976
Zhu Y, Chen H, Fan J, Wang Y, Li Y, Chen J, Fan J, Yang S, Hu L, Leung H, Mew TW, Teng PS, Wang Z, Mundt CC (2000) Genetic diversity and disease control in rice. Nature 406:718–722
Zhu Z, Huo Y, Wang X, Huang J, Wu X (2004) Analysis of simple sequence repeats markers derived from Phytophthora sojae expressed sequence tags. Chin Sci Bull 49(19):2041–2046
Zou J, Lee J, Singh R, Xu SS, Cregan PB, Hymowitz T (2003) Assignment of molecular linkage groups to the soybean chromosomes by primary trisomics. Theor Appl Genet 107:745–750
Acknowledgments
This study was conducted in the Key Laboratory of Soybean Biology of Chinese Education Ministry and Soybean Development Centre of Agricultural Ministry, financially supported by National High Technology Project (2006AA10Z1F1 and 2006AA100104-4), 948 project of Agricultural Ministry of China (2006-G5), National Nature Science Foundation projects (30971810, 60932008), National 973 Project (2009CB118400), Technology project of Education Ministry of Heilongjiang Province (11541025).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Lightfoot.
X. Li and Y. Han have equal contributions to the paper.
Rights and permissions
About this article
Cite this article
Li, X., Han, Y., Teng, W. et al. Pyramided QTL underlying tolerance to Phytophthora root rot in mega-environments from soybean cultivars ‘Conrad’ and ‘Hefeng 25’. Theor Appl Genet 121, 651–658 (2010). https://doi.org/10.1007/s00122-010-1337-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1337-2