Abstract
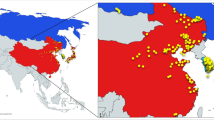
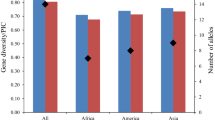
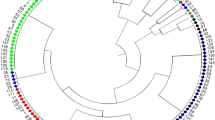
Annual wild soybean (Glycine soja Sieb. et Zucc.) is believed to be a potential gene source for future soybean improvement in coping with the world climate change for food security. To evaluate the wild soybean genetic diversity and differentiation, we analyzed allelic profiles at 60 simple-sequence repeat (SSR) loci and variation of eight morph-biological traits of a representative sample with 196 accessions from the natural growing area in China. For comparison, a representative sample with 200 landraces of Chinese cultivated soybean was included in this study. The SSR loci produced 1,067 alleles (17.8 per locus) with a mean gene diversity of 0.857 in the wild sample, which indicated the genetic diversity of G. soja was much higher than that of its cultivated counterpart (total 826 alleles, 13.7 per locus, mean gene diversity 0.727). After domestication, the genetic diversity of the cultigens decreased, with its 65.5% alleles inherited from the wild soybean, while 34.5% alleles newly emerged. AMOVA analysis showed that significant variance did exist among Northeast China, Huang-Huai-Hai Valleys and Southern China subpopulations. UPGMA cluster analysis indicated very significant association between the geographic grouping and genetic clustering, which demonstrated the geographic differentiation of the wild population had its relevant genetic bases. In comparison with the other two subpopulations, the Southern China subpopulation showed the highest allelic richness, diversity index and largest number of specific-present alleles, which suggests Southern China should be the major center of diversity for annual wild soybean.


Similar content being viewed by others
References
Abe J, Xu DH, Suzuki Y, Kanazawa A (2003) Soybean germplasm pools in Asia revealed by nuclear SSR. Theor Appl Genet 106:445–453
Buckler ESIV, Thornsberry JM, Kresovich S (2001) Molecular diversity and domestication of grasses. Genet Res 77:213–218
Carter TE, Nelson RL, Sheller CH, Cui ZL (2004) Genetic diversity in soybean. In: Boerma HR, James ES (eds) Soybeans: improvement, production, and uses, 3rd edn. ASA,CSSA and SSSA, Madison, pp 309–310
Chakrabortry R, Jin L (1993) A unified approach to study hypervariable polymorphisms: statistical considerations of determining relatedness and population distances. In: Pena SDJ, Chakraborjt R, Epplen JT, Jeffreys AJ (eds) DNA fingerprinting: state of the science. Birkhäuser Verlag, Basel, pp 153–175
Chen YW, Nelson RL (2004) Genetic variation and relationships among cultivated, wild, and semiwild soybean. Crop Sci 44:316–325
Chen YW, Nelson RL (2005) Relationship between origin and genetic diversity in Chinese soybean germplasm. Crop Sci 45:1645–1652
Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai TT, Lohnes DG, Chung J, Especht J (1999) An intergrated genetic linkage map of the soybean genome. Crop Sci 39:1464–1490
Diwan N, Cregan PB (1997) Automated sizing of fluorescent-labeled simple sequence repeat (SSR) markers to assay genetic variation in soybean. Theor Appl Genet 95:723–733
Dong YS, Zhuang BC, Zhao LM, Sun H, He MY (2001) The genetic diversity of annual wild soybeans grown in China. Theor Appl Genet 103:98–1030
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Excoffier L, Schneider S (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolut Bioinform Online 1:47–50
Excoffier L, Smouse P, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes:application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fujita R, Ohara M, Okazaki K, Shimamoto Y (1997) The extent of natural cross-pollination in wild soybean (Glycine soja). J Hered 88:124–128
Gai JY, Xu DH, Gao Z, Shimamoto Y, Abe J, Fukushi H, Kitajima S (2000) Studies on the evolutionary relationship among eco-types of G. max and G. soja in China. Acta Agron Sin 26(5):513–520
Kiang YT, Chiang YC, Kaizuma N (1992) Genetic diversity in natural populations of wild soybean in Iwate prefecture, Japan. J Hered 83:325–329
Kuroda Y, Kaga A, Tomooka N, Vaughan AD (2006) Population genetic structure of Japanese wild soybean(Glycine soja) based on microsatellite variation. Mol Ecol 15:959–974
Li FS (1993) Studies on the ecological geographical distribution of the Chinese resources of wild soybean. Sci Agr Sin 26:47–55
Li Zl, Nelson RL (2002) RAPD marker diversity among cultivated and wild soybean accessions from four Chinese Provinces. Crop Sci 42:1737–1744
Li J, Tao Y, Zheng SZ, Zhou JL (1995) Isozymatic differentiation in local population of Glycine soja sieb & zucc. Acta Bot Sin 37:669–676
Liu K, Muse SV (2005) PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21:2128–2129
Maughan PJ, Saghai MA, Buss GR (1995) Microsatellite and amplified sequence length polymorphisms in cultivated and wild soybean. Genome 38:715–723
Michalakis Y, Excoffier L (1996) A generic estimation of population subdivision using distances between alleles with special reference to microsatellite loci. Genetics 142:1061–1064
Narvel JM, Fehr WR, Chu WC, Grant D, Shoemaker RC (2000) Simple sequence repeat diversity among soybean plant introductions and elite genotypes. Crop Sci 40:1452–1458
Pei YL, Wang L, Ge S, Wang LZ (1996) Studies on genetic diversity of Glycine soja isozyme variation in four populations. Soybean Sci 15:302–309
Powell W, Morgante M, Doyle JJ, McNicol JW, Tingey SV, Rafalski AJ (1996) Genepool variation in Genus Glycine subgenus Soja revealed by polymorphic nuclear and chloroplast microsatellites. Genetics 144:791–803
Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128
Tozuka A, Fukushi H, Hirata T, Ohara M, Kanazawa A, Mikaml T, Abe J, Shimamoto Y (1998) Composite and clinal distribution of Glycine soja Japan revealed by RFLP analysis of mitochondrial DNA. Theor Appl Genet 96:170–176
Wang KJ, Takahata Y (2007) A preliminary comparative evaluation of genetic diversity between Chinese and Japanese wild soybean (Glycine soja) germplasm pools using SSR markers. Genet Resour Crop Evol 54:157–165
Wang LX, Guan RX, Liu ZX, Chang RZ, Qiu LJ (2006) Genetic diversity of chinese cultivated soybean revealed by SSR markers. Crop Sci 46:1032–1038
Xu DH, Gao Z, Tian QZ, Gai JY, Fukushi H, Kitajma S, Abe J, Shimamoto Y (1999) Genetic diversity of the annual soybean (Glycine soja) in China. Appl Environ Biol 5:439–443
Xu DH, Abe J, Gai JY, Shimamoto Y (2002) Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theor Appl Genet 105:645–653
Zhuang BC, Xu H, Wang YM (1996) Polymorphism and geographical distribution of the stem and leaf characters of wild soybean (Glycine soja) in China. Acta Agron Sin 22:583–586
Acknowledgments
The project was supported by the Natural Science Foundation of China (30671266), the National Key Basic Research Program (2006CB101708, 2009CB118404), the National “863” Program (2006AA100104) and the MOE 111 Project (B08025). The authors would thank the editor and reviewers for their relevant comments on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by A. Schulman.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wen, Z., Ding, Y., Zhao, T. et al. Genetic diversity and peculiarity of annual wild soybean (G. soja Sieb. et Zucc.) from various eco-regions in China. Theor Appl Genet 119, 371–381 (2009). https://doi.org/10.1007/s00122-009-1045-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1045-y