Abstract
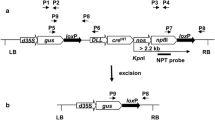
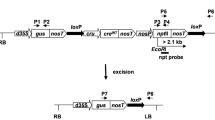
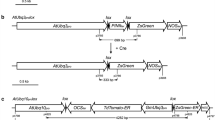
This work is focused on the generation of selectable marker-free transgenic tobacco plants using the self excision Cre/loxP system that is controlled by a strong seed specific Arabidopsis cruciferin C (CRUC) promoter. It involves Agrobacterium-mediated transformation using a binary vector containing the gus reporter gene and one pair of the loxP sites flanking the cre recombinase and selectable nptII marker genes (floxed DNA). Surprisingly, an ectopic activation of CRUC resulting in partial excision of floxed DNA was observed during regeneration of transformed cells already in calli. The regenerated T0 plants were chimeric, but no ongoing ectopic expression was observed in these one-year-long invitro maintained plants. The process of the nptII removal was expected in the seeds; however, none of the analysed T0 transgenic lines generated whole progeny sensitive to kanamycin. Detailed analyses of progeny of selected T0-30 line showed that 10.2% GUS positive plants had completely removed nptII gene while the remaining 86.4% were still chimeras. Repeated activation of the cre gene in T2 seeds resulted in increased rate of marker-free plants, whereas four out of ten analysed chimeric T1 plants generated completely marker-free progenies. This work points out the feasibility as well as limits of the CRUC promoter in the Cre/loxP strategy.





Similar content being viewed by others
References
Becerra C, Puigdomenech P, Vicient CM (2006) Computational and experimental analysis identifies Arabidopsis genes specifically expressed during early seed development. BMC Genomics 7:38. doi:10.1186/1471-2164-7-38
Belostotsky DA, Meagher RB (1996) A pollen-, ovule-, and early embryo-specific poly(A) binding protein from Arabidopsis complements essential functions in yeast. Plant Cell 8:1261–1275
Bevan MW, Flavell RB, Chilton MD (1983) A chimeric antibiotic resistance gene as a selection marker for plant cell transformation. Nature 304:184–187
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantise of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Coppoolse ER, deVroomen MJ, Roelofs D, Smit J, van Gennip F, Hersmus BJM, Nijkamp JJ, van Haaren MJ (2003) Cre recombinase expression can result in phenotypic aberrations in plants. Plant Mol Biol 51:263–279
Cuellar W, Gaudin A, Solórzano D, Casas A, Ñopo L, Chudalayandi P, Medrano G, Kreuze J, Ghislain M (2006) Self-excision of the antibiotic resistance gene nptII using a heat inducible Cre/loxP system from transgenic potato. Plant Mol Biol 62:71–82
Hoff T, Schnorr KM, Mundy J (2001) A recombinase-mediated transcriptional induction system in transgenic plants. Plant Mol Biol 45:41–49
Höglund AS, Rödin J, Larsson E, Rask L (1992) Distribution of napin and cruciferin in developing rape seed embryos. Plant Physiol 98:509–515
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusion: β-glucuronidase as sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3908
Keenan RJ, Stemmer WP (2002) Nontransgenic crops from transgenic plants. Nat Biotechnol 20:215–216
Li Z, Xing A, Moon BP, Burgoyne SA, Guida AD, Liang H, Lee C, Caster CS, Barton JE, Klein TM, Falco SC (2007) A Cre/loxP-mediated self-activating gene excision system to produce marker free transgenic soybean plants. Plant Mol Biol 65:329–341
Liu HK, Ch Yang, Wei ZM (2005) Heat shock-regulated site-specific excision of extraneous DNA in transgenic plants. Plant Sci 168:997–1003
Luo K, Duan H, Zhao D, Zheng X, Deng W, Chen Y, Stewart CN, McAvoy R, Jiang X, Wu Y, He A, Pei Y, Li Y (2007) ‘GM-gene-deletor’: fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or CRE recombinase on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol J 5:263–274
Marjanac G, De Paepe A, Peck I, Jacobs A, De Buck S, Depicker A (2008) Evaluation of CRE-mediated excision approaches in Arabidopsis thaliana. Transgenic Res 17:239–250
Mlynárová L, Loonen A, Heldens J, Jansen RC, Keizer P, Stiekema WJ, Nap JP (1994) Reduced position effect in mature transgenic plants conferred by the chicken lysozyme matrix associated region. Plant Cell 6:417–426
Mlynárová L, Nap JP (2003) A self-excising Cre recombinase allows efficient recombination of multiple ectopic heterospecific lox sites in transgenic tobacco. Transgenic Res 12:45–57
Mlynárová L, Conner AJ, Nap JP (2006) Directed microspore-specific recombination of transgenic alleles to prevent pollen-mediated transmission of transgenes. Plant Biotechnol J 4:445–452
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:2002–2007
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann J (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37:501–506
Stuitje AR, Verbree EC, van der Linden KH, Mietkiewska E, Nap JP, Kneppers TJA (2003) Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol J 1:301–309
Suzuki M, Kao CY, Cocciolone S, McCarty DR (2001) Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J 28:409–418
Trindade LM, Horvath B, Bachem C, Jacobsen E, Visser RGF (2003) Isolation and functional characterization of a stolon specific promoter from potato (Solanum tuberosum L.). Gene 303:77–87
Vaculková E, Moravčíková J, Matušíková I, Bauer M, Libantová J (2007) A modified low copy number binary vector pUN for Agrobacterium-mediated transformation. Biol Plant 51:538–540
van Engelen A, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4:288–290
Verweire D, Verleyen K, De Buck S, Claeys M, Angenon G (2007) Marker-free transgenic plants through genetically programmed auto-excision. Plant Physiol 145:1220–1231
Zhang Y, Li H, Ouyang B, Lu Y, Ye Z (2006) Chemical-induced auto-excision of selectable markers in elite tomato plants transformed with a gene conferring resistance to lepidopteran insects. Biotechnol Lett 28:1247–1253
Zheng X, Deng W, Luo K, Duan H, Chen Y, McAvoy R, Song S, Pei Y, Li Y (2007) The cauliflower mosaic virus (CaMV) 35S promoter sequence alters the level and patterns of activity of adjacent tissue- and organ-specific gene promoters. Plant Cell Rep 26:1195–1203
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632
Acknowledgments
We thank Drs. Mlynárová and Nap for providing plasmids pFLUAR101, pLM91-containing CRUC-cre expression unit and for helpful discussion. We thank Dr. Salaj for help with microscopic techniques, and Dr. Matušíková for her kind proof reading of the manuscript. The authors thank Anna Fábelová for invitro plant care. The study was funded by Scientific Grant Agency of the Ministry of Education of Slovak Republic and Slovak Academy of Sciences 2/0011/08.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. F. Quiros.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2008_866_MOESM1_ESM.tif
Electronic Supplementary Material S1 Examples of Southern blot analysis of parental T0 plants. a VspI-digested DNA probed with the GUS specific probe. The bands of a 2.9 kb correspond to the internal gus fragment. All additional bands (>1.5 kb) correspond to the border fragments and indicate the number of independent transgene copies. The absence of the internal 2.9 kb gus fragment in the plant T0-8 indicates an incomplete gus gene integration. b EcoRI-digested DNA probed with the NPTII specific probe. The bands (>2.1 kb) correspond to the number of independent transgene copies. NT – non-transgenic control plant, asterisks indicate single copy transformants used in further analyses. (TIFF 9924 kb)
122_2008_866_MOESM2_ESM.tif
Electronic Supplementary Material S2 PCR detection of premature excision in T0 plants. Photographs of ethidium bromide-stained 1% agarose gels carrying PCR products obtained from the single-copy T0 tobacco plants. a PCR with the primers P7/P6 that amplified a 0.59 kb fragment indicating excision of the nptII gene (NPT-). b PCR with the primers P5/P6 that amplified a 1.8 kb fragment corresponding to the nptII gene (NPT+). c PCR products obtained on the same plant set with the actin primers. The lane M contains 1 kb DNA ladder (Fermentas) as a size marker, next follow the PCR products of individual T0 plants, the last lane represents PCR product of plasmid pEV2 (EV2). (TIFF 6356 kb)
122_2008_866_MOESM3_ESM.tif
Electronic Supplementary Material S3 PCR analyses of premature excision in developing transgenic calli. Genomic DNA from transgenic calli was subjected to PCR analysis with the primers P7/P6 that amplified a 0.59 kb PCR fragment indicating the nptII gene excision (NPT-) and the primers P5/P6 that amplified a 1.8 kb fragment corresponding to the nptII gene (NPT+). The actin primers were used as a PCR control. The lane M contains 1 kb DNA ladder (Fermentas) as a size marker, C – transgenic calli, NT – non-transformed control, EV2 – plasmid pEV2. (TIFF 3590 kb)
Rights and permissions
About this article
Cite this article
Moravčíková, J., Vaculková, E., Bauer, M. et al. Feasibility of the seed specific cruciferin C promoter in the self excision Cre/loxP strategy focused on generation of marker-free transgenic plants. Theor Appl Genet 117, 1325–1334 (2008). https://doi.org/10.1007/s00122-008-0866-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0866-4