Abstract
The genes encoding for 18S–5.8S–28S ribosomal RNA (rDNA) are both conserved and diversified. We used rDNA as probe in the fluorescent in situ hybridization (rDNA-FISH) to localized rDNAs on chromosomes of 15 accessions representing ten Oryza species. These included cultivated and wild species of rice, and four of them are tetraploids. Our results reveal polymorphism in the number of rDNA loci, in the number of rDNA repeats, and in their chromosomal positions among Oryza species. The numbers of rDNA loci varies from one to eight among Oryza species. The rDNA locus located at the end of the short arm of chromosome 9 is conserved among the genus Oryza. The rDNA locus at the end of the short arm of chromosome 10 was lost in some of the accessions. In this study, we report two genome specific rDNA loci in the genus Oryza. One is specific to the BB genome, which was localized at the end of the short arm of chromosome 4. Another may be specific to the CC genome, which was localized in the proximal region of the short arm of chromosome 5. A particular rDNA locus was detected as stretched chromatin with bright signals at the proximal region of the short arm of chromosome 4 in O. grandiglumis by rDNA-FISH. We suggest that chromosomal inversion and the amplification and transposition of rDNA might occur during Oryza species evolution. The possible mechanisms of cyto-evolution in tetraploid Oryza species are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genes encoding for 18S–5.8S–28S ribosomal RNA (rDNA) occur as tandem arrays at one or several specific regions on chromosomes. In higher eukaryotes, each rDNA repetitive unit contains a transcriptional unit, including the genes for 18S–5.8S–28S and two internal transcribed spacers (ITSs), and an intervening intergenic spacer (IGS) region. These genes are highly conserved among various organisms although the spacer regions are less conserved than the genes themselves (Weider et al. 2005). The chromosomal segment harboring these genes is known as a nucleolar organizing region (NOR), which is often associated with a nucleolus. The rDNAs could change rapidly both in copy number and chromosomal location in plant genomes (Schubert and Wobus 1985; Raina and Mukai 1999). The number and location of NORs are varied among related genomes; therefore, NOR chromosome could be used as a chromosomal landmark to provide valuable evidence regarding genome evolution at chromosomal and molecular levels. The fluorescent in situ hybridization method with rDNA probe (rDNA-FISH) is more advantageous than the classical silver-staining analysis because it can detect rDNAs irrespective of their transcriptional activity (Linde-Laursen et al. 1992; Chung et al. 1993). Besides, the mass and intensity of the resulting fluorescent signal could reflect the copy number of genes arrayed at that site (Cerbah et al. 1998). The polymorphism of rDNAs in copy number of repeats and the chromosomal position are visible and comparable in the same experiment and have been investigated in several plant genomes (Leitch and Heslop-Harrison 1992; Linde-Laursen et al. 1992; Raina and Mukai 1999; Li and Zhang 2002). Those results have provided valuable information of genomic organizations at the chromosomal level.
Cultivated rice (Oryza sativa, L) is one of the most important staple food crops in the world. The genus Oryza comprises about 24 species, including two cultivated species, O. sativa and O. glaberrima, and wild species. The Oryza species, including diploids (2n = 24) and tetraploids (2n = 48), are classified genetically into ten genome types, i.e. AA, BB, CC, BBCC, CCDD, EE, FF, GG, JJHH and JJKK genomes (Vaughan et al. 2003). The phylogenetic relationships in the genus Oryza have been studied broadly through various molecular markers (Ge et al. 1999). Furthermore, in silico analysis through the effective physical mapping of DNA markers will allow for detailed comparative genome analyses of these species. In those studies, chromosomal duplications and rearrangements have been reported (Wang et al. 2005; Yu et al. 2005). However, rDNA-FISH can provide an alternative and efficient solution to investigate cyto-evolution at the chromosome level among Oryza species.
Oryza sativa ssp. japonica has a NOR at the end of the short arm of chromosome 9 (Kurata and Omura 1978; Fukui and Iijima 1991; Chung et al. 1993) and O. sativa ssp. indica has one more NOR at the end of the short arm of chromosome 10 (Chung and Wu 1987; Chung et al. 1993). To date, the chromosomal locations of rDNAs in several cultivars and the wild species of rice have been analyzed using the FISH technique (Ohmido and Fukui 1995; Shishido et al. 2000). In this study, we investigated the chromosomal locations of the rDNA loci in 15 accessions including cultivated rice and the wild species of rice by FISH. Our results show that rDNA genes are polymorphic in the number of loci, in the copy number of the repeat units, and in chromosomal positions among the Oryza species. Based on these results the possible mechanisms of cyto-evolution in Oryza species are discussed.
Materials and methods
Plant material
The rice species and cultivars used in this study are listed in Table 1. The original seeds of the wild species of rice were kindly provided by the late Professor H. I. Oka, National Institute of Genetics, Japan (http://www.pgcdna.co.jp/cgi-bin/wrdb/content.cgi). In addition to those wild species of rice, the following four cultivars were used in this study. Nipponbare (O. sativa ssp. japonica), a well-known cultivar, was used as the representative genome in the whole genome sequencing project (International Rice Genome Sequencing Project 2005). TNG 67 is one the most popular japonica type cultivars in Taiwan for its high yield potential and environmental adaptation and tolerance to blast (http://www.tari.gov.tw). IR36 is one of well-known cultivars (O. sativa ssp. indica) released by the Internal Rice Research Institute (IRRI) with high yield and photo-insensitive characters. Dular is a medium-grain cultivar from India.
DNA probe preparation
Plasmid pTA71 which contains approximately 9 kb of the 18S–5.8S–26S rRNA repeat units of Triticum aestivum (Gerlach and Bedbrook 1979) was used in this study. Plasmid pRCS2 that contains rice centromere specific CentO repeats (Dong et al. 1998) was used to indicate the centromere position on the rice chromosomes. Purified plasmid DNA was labeled with biotin-14-dATP or digoxigenin-11-dUTP by nick-translation (Roche Diagnostics GmbH, Penzberg, Germany) and then used as FISH probes.
Chromosome preparation and fluorescent in situ hybridization
Chromosome preparation and the FISH procedure followed the previously described protocols (Kao et al. 2006). Young and healthy root tips were harvested, pretreated in 2 mM 8-hydroxyquinoline at 25°C for 2 h to achieve prometaphase accumulation, and fixed in freshly prepared Farmer’s fluid (95% ethanol:glacial acetic acid = 3:1). Root tips were macerated with 6% cellulose (Onoauka R-10, Yakukt Honsha, Japan) and 6% pectinase (Sigma Chemical Co., St. Louis, MO.) in 75 mM KCl, pH 4.0 at 37°C for 75 min then squashed on a slide with the same fixative. Slides were air-dried and stored at −80°C until required.
Slides were dehydrated through an ethanol series (70, 95, and 100%, 2 min each) prior to be used in fluorescence in situ hybridization. After baking on 60°C hotplate for 30 min, the chromosome preparations were pretreated with pepsin (1–2 mg/mL, in 10 mM HCl) at 37°C for 15 min. The chromosomes were denatured in 70% deionized formamide (in 2× SSC) at 80°C for 90 s, and immediately immersed into ice-cold 70, 95 and 100% ethanol sequentially and air-dried. The hybridization mixture contained 50% deionized formamide, 2× SSC, sheared salmon sperm DNA (1 μg/μL), 10% dextran sulfate, and probe DNA (10–100 ng in 20 μL, per slide). Hybridization was carried out in a moisture box at 37°C for overnight. After hybridization, slides were washed in 2× SSC at room temperature for 5 min, at 42°C for 10 min, and then for another 5 min at room temperature. Hybridization sites of biotin- or digoxigenin-labeled probes were immunologically detected using fluorescein isothiocyanate (FITC)-conjugated avidin (Vector Laboratories, CA, USA) or a rhodamine-conjugated anti-digoxigenin antibody (Roche Diagnostics GmbH, Penzberg, Germany), respectively. Chromosomes were counterstained with DAPI in an antifade solution (Vector Laboratories, CA, USA). Digital images were captured by a CCD camera (Cool Snapfx, Photometrics, Tucson, AZ, USA) driven by Image-Pro Plus software (version 4.5.1, Media Cybernetics, Yorktown, VA, USA) through an epifluorescent microscope (Axioplan, Carl Zeiss AG, Germany). Final image was edited with Adobe Photoshop 6.0 (Adobe Systems Incorporated, San Jose, CA, USA).
Results
Chromosome identification and karyotyping
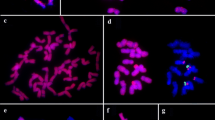
Each rice chromosome can be identified based on length and morphology at somatic prometaphase and meiotic pachytene stage. Twelve pairs of rice chromosomes are numbered according to their length in descending order. The chromosome complements (Fig. 1a) and a standard karyotype (Fig. 1b) of rice, O. sativa ssp. indica cv. IR36, at somatic prometaphase are shown in Fig. 1. By conventional Giemsa staining, each pair of rice chromosome presents distinguishable features, such as relative length, centromere position (arm ratio), and heterochromatin distribution pattern. Both chromosomes 9 and 10 usually attached with nucleolus are nucleolar chromosomes. In this study, chromosome identification is based on this established karyotype of IR36 and previously reported (Chen and Wu 1982). To simplify, the numbers of rDNA loci per respective haploid genome are mentioned in the following sections.
Genomes with only one rDNA locus
Oryza sativa ssp. japonica cv. Nipponbare (AA), O. glaberrima (AA), and O. australiensis (EE) have only one rDNA locus at the end of the short arm of chromosome 9 in respective genome (Fig. 2a–c). The fluorescent signals of rDNA-FISH revealed that the NOR appeared as two extensions from condensed sister chromatids at prometaphase.
One or two pairs of rDNA loci (red) were mapped by FISH on somatic prometaphase chromosomes (blue) of a O. sativa ssp. japonica cv. Nipponbare (AA), b O. glaberrima (AA), and c O. australiensis (EE), d O. sativa ssp. japonica cv. TNG 67, e O. sativa ssp. indica cv. IR36 and f Dular, g O. rufipogon (W0107), and h O. rufipogon (W1623). Note: the signals on chromosome 10 (arrows) were apparently less than those on chromosome 9 (arrowheads) in d and e. Centromeres are indicated by biotin-labeled pRCS2 (green) in a–c and h. Chromosomes were counterstained with DAPI (blue) and identified based on their morphology and length. Scale bar 10 μm
Genomes with two rDNA loci
In addition to the rDNAs on chromosome 9, the second site was found at the end of the short arm of chromosome 10 in several species with AA genome, including O. sativa ssp. japonica cv. TNG 67, indica cv. IR36 and Dular, and O. rufipogon (W0107, W1623) (Fig. 2d–h). In TNG 67 and IR36, bright fluorescent signals of rDNA-FISH presented as extensions from the ends of the short arms of chromosome 9 (Fig. 2d, e). The fluorescent signals of rDNA-FISH could reflect the relative length of rDNA arrays at that position, therefore, the rDNA repeats at the end of the short arm of the chromosome 10 are less than those at chromosome 9 in IR36 and TNG 67; however, the length of rDNA arrays on both sites were similar in the other accessions (Fig. 2f–h).
Genomes with three rDNA loci
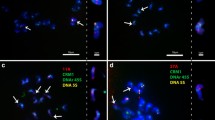
Oryza punctata (W1593 and W1577) with BB genome and O. officinalis with CC genome have three rDNA loci, respectively. In addition to those two loci on chromosomes 9 and 10, the third locus was detected at the end of the short arm of chromosome 4 in O. punctata (BB; Fig. 3a–b), which was first reported in BB genome. Among rice chromosome complements, chromosome 4 can be easily identified by its relative short and heterochromatic short arm (Fig 1b). In O. officinalis (CC), the third locus was detected in the proximal region at the short arm of chromosome 5 (Fig. 3c), which is the newly identified rDNA locus in rice genomes. Chromosome 5 can be identified as a submetacentric chromosome (Kao et al. 2006), which is a little shorter than chromosome 4 (Fig. 1b).
FISH localization of rDNA (red) loci on somatic prometaphase chromosomes (blue) of a O. punctata (BB, W1593), b O. punctata (BB, W1577), c O. officinalis (CC), d O. puntata (BBCC), and e O. minuta (BBCC). For each accession, chromosome complements with rDNA-FISH signals were shown at left and respective karyotypes were arranged and shown at right. Scale bar 10 μm
rDNA loci in tetraploid rice species
Tetraploid O. punctata with BBCC genome had six rDNA loci in its haploid genome (Fig. 3d). The 45S rDNAs were detected at the ends of the short arm of two pairs of chromosome 9 (9B and 9C) and chromosome 10 (10B and 10C), one pair of chromosome 4 (4B), and the sixth site at the proximal region of the short arm of one pair of chromosome 5 (5C). However, another tetraploid, O. minuta with BBCC genome had four to five rDNA loci in its haploid genome (Fig. 3e). Four rDNA loci were consistently detected at the end of the short arm of two pairs of chromosome 9 (9B and 9C) and chromosome 10 (10B and 10C), respectively. The fifth rDNA locus was observed at the end of the short arm of one pair of chromosome 4 in some nuclei.
Oryza latifolia, which is a tetraploid with CCDD genome had four rDNA loci on the end of the short arm of two homologous pairs of chromosome 9 (Fig. 4a–b). However, another tetraploid O. grandiglumis with CCDD genome had eight rDNA loci in its haploid genome. These loci were localized at the end of the short arm of two homologous pairs of chromosomes 9 and 10, and in the proximal region at the short arms of two homologous pairs of chromosomes 4 and 5, respectively (Fig. 5c). The rDNA locus next to the centromere of chromosome 4 is the most particular among all the rDNA sites detected by rDNA-FISH in rice species. The stretched rDNA arrays on one of the homologous chromosome 4 were labeled with bright fluorescent signals of rDNA-FISH. The rDNA arrays at the other chromosome 4 seemed shorter than that at the former.
FISH localization of rDNA (red) loci on somatic prometaphase chromosomes (blue) of tetraploid Oryza species with the CCDD genome. Two pairs of rDNAs were mapped on chromosome 9 of O. latifolia with different accession numbers, a W0019 and b W0542, respectively. c Eight pairs of rDNAs were mapped on the chromosomes of O. gradiglumis including two homologous pairs of chromosomes 4, 5, 9, and 10. The intercalary rDNA sites on chromosome 4 were labeled with strong and bright fluorescent signals (arrows). For each accession, chromosome complements with rDNA-FISH signals were shown at left and respective karyotypes were arranged and shown at right. Scale bar 10 μm
Various transcriptional activities among rDNA loci
In all accessions, the rDNAs arrayed at the ends of the short arm of chromosome 9 were detected as two extensions from relatively condensed prometaphase chromosomes. In some accessions, such as O. rufipogon (Fig. 2g–h), and O. officinalis (Fig 3c), rDNA arrays at the end of the short arm of chromosome 10 also appeared as stretched chromatin feature as that on the chromosome 9. The rDNA-FISH signals at chromosome 4, except one in O. grandiglumis (Fig 5c), or at chromosome 5 usually appeared as dots or bands, never as stretched chromatin. These results suggest that rDNA loci may vary in length of rDNA arrays. Besides, we observed numerous small nucleular-like globules attached with pachytene chromosomes (Fig. 5a, b). We suggest that these may be the products of those “minor” rDNA loci with relative short rDNA arrays, which may be beyond the limitation of FISH resolution in rice genomes.
Discussion
Owing to the improvements in resolution and sensitivity of FISH techniques, more and more new rDNA loci have been identified in several cases. In this study, we used an rDNA repetitive unit cloned from wheat (Gerlach and Bedbrook 1979) as probe in FISH experiments. As mentioned above, this repetitive unit contains a highly conserved transcription region of the 18S–5.8S–26S rRNA gene including the internal transcribed spacer (ITS), but excluding the intergenic spacer (IGS), which has considerable divergence from the ancestral sequence. However, comparison of the IGS of rDNAs, revealed several highly conserved regions among cereals including rice, maize, wheat, and rye (Cordesse et al. 1993). Furthermore, the length polymorphism is responsible for the variations of IGS regions among Oryza species (Sano and Sano 1990). Although sequences in ITSs may be less conserved, the highly conserved transcriptional regions made this clone as a useful probe for rDNA-FISH in several species including Sorghum species (Sang and Liang 2000), rice (Cheng et al. 2001b), and Arabidopsis thaliana (Murata et al. 2006).
Here we report two genome specific rDNA loci in the genus Oryza and reveal polymorphism of rDNA loci in Oryza species including diploids and tetraploids through rDNA-FISH technique. Our rDNA-FISH results and minor nucleoli observed at pachytene stage (Fig 5a, b) led us to suggest that, in addition to the major rDNAs at the short arm end of chromosomes 9 and 10, there are numerous minor rDNA loci in common progenitor genome of Oryza species, some of these minor rDNA loci might accumulate more rDNAs via unequal crossing over or retrotransposition and finally became genome specific and rDNA-FISH detectable chromosome landmarks. Similarly, rDNA sites might be lost during evolution.
Chromosomal rearrangements, such as translocation or inversion, involving the segments bearing the rDNAs would shift the NORs location as reported in Psathyrostachys (Orgaard and Heslop-Harrison 1994) and in Allium fistulosum (Ricroch et al. 1992). However, previous studies have shown high karyotypic similarity in Oryza species (Chen and Wu 1982), which suggested that chromosomal rearrangements on a large scale are not frequent in the genus Oryza. The variation of rDNAs in the Oryza species may be a result of transposon mobility and amplification of cryptic minor rDNA sites by unequal crossing over rather than from chromosomal rearrangements. For example, deletion must have occurred in rDNA arrays at the end of the short arm of chromosome 10 in some Oryza species. O. rufipogon which is considered to be the wild progenitor of Asian rice O. sativa has two rDNA sites (Fig. 2h, Fukui et al. 1994). The different number of rDNA locus between japonica rice and indica rice means japonica rice lost the rDNA site on chromosome 10 after the differentiation between japonica rice and indica rice. Nevertheless, TNG 67 which is a japonica type cultivar (Fig. 2d) possibly gained the second rDNA array at the short arm end of chromosome 10 from its indica type rice ancestral parent by unequal crossing over; while indica type cultivar IR36 (Fig. 2e) lost a part of DNAs array on this site by the same mechanism during the breeding process. African cultivated rice O. glaberrima and O. rufipogon were grouped into two separate clusters based on the earlier observations (Aggarwal et al. 1999), as well as based on the rDNA-FISH results presented in a previous report (Fukui et al. 1994; Ohmido and Fukui 1995) and in this study (Fig. 2b, g, h). Although O. latifoia had been detected to have five rDNA loci by Fukui et al. (1994), we suggest that O. latifolia (CCDD, Fig. 4a, b) is similar to O. australiensis (W0008, EE, Fig. 2c) in rDNA distribution. These results might provide one more piece of evidence for supporting the proposition that the EE genome species is closely related to the DD genome progenitor that gave rise to the CCDD genome (Ge et al. 1999). O. brachyantha (FF genome) which is distantly related to the species containing AA, BB and CC genomes and to the EE genome species (Ge et al. 1999) has only one rDNA locus at the end of the short arm of chromosome 9 (Shishido et al. 2000). These results suggested that the rDNA locus at the end of the short arm of chromosome 10 might appear after the clade containing AA, BB, and CC genomes was separated from other clades.
The rDNA site at the end of the short arm of chromosome 4 was first reported in CC genome species, O. officinalis and O. eichingeri by Shishido et al (2000). However, we detected this rDNA site in species with BB genome, including O. punctata (BB, Fig. 3a, b), O. punctata (BBCC, Fig. 3d ) and O. minuta (BBCC, Fig. 3e) using the FISH technique. We suggest that rDNAs at the end of the short arm of chromosome 4 may be specific to the Oryza species with the BB genome.
In this study, we report the newest rDNA locus in the proximal region at the short arm of chromosome 5. This intercalary rDNA site seems specific to the Oryza species with the CC genome, including O. officinalis (CC, Fig. 3c), O. punctata (BBCC, Fig. 3d), and O. grandiglumis (CCDD, Fig. 5c). However, no detectable intercalary rDNAs were found in O. minuta (BBCC, Fig. 3e) and O. latifolia (CCDD, Fig. 4a, b), although both have CC genomes. It suggests that genomic modification had occurred during the formation of these allotetraploids.
Two parental genomes in an allopolyploid may possibly undergo genomic alterations, such as recombination and transposon reactivation, to achieve genomic stability (for review see: Comai 2000; Comai et al. 2003). Our results suggested that a few alterations including deletion, amplification, and chromosomal rearrangements have occurred in allotetraploid rice genomes. It also implies that those two BBCC genomes, O. punctata and O. minuta, may have had different origins. Apart from the ancestral origin of the DD genome, which is still a mystery, rDNAs on chromosome 5 and chromosome 10 inherited from the CC genome were lost in O. latifolia (CCDD).
The number and distribution of the rDNA loci in O. grandiglumis (CCDD) genome demonstrate that a series of genome modification events had occurred during its formation. Its ancestral parents, especially the presumed diploid descendant with the DD genome, were not available, as possible genomic alterations are hard to trace. Based on our results of rDNA-FISH, we suggest, homologous pairing and consequent recombination and rDNAs transposition may have happened prior to the hybrid genome doubling to become an allotetraploid and genomic stability being achieved. For example, the rDNA sites at the short arm end of chromosomes 5 and 10 from C genome were transferred to chromosome 5 from D genome via homologous pairing and unequal crossing over. The intercalary rDNA site on chromosome 4 in O. grandiglumis (CCDD genome, Fig 4c) implied that at least two events have occurred during the evolution of the genus Oryza. First, genes may have been amplified in the rDNA arrays at this site, which might have been inherited from its ancestral progenitor with minor rDNAs at this site. The gene amplification might have resulted from the formation of allotetraploid. Second, chromosomal inversion occurred in the segment involving the short arm of chromosome 4 with rDNAs. This shifted the rDNAs from the end of the short arm to the region next to the centromere of chromosome 4. Morphologically, the short arm of rice chromosome 4 appears as a highly condensed heterochromatin at metaphase, implying that this region is composed of highly repetitive sequence. As shown in previous studies (Feng et al. 2002), chromosome 4 of O. sativa contained abundant and various types of repetitive sequences, accounting for 18.2% of decoded chromosome 4, and these repetitive sequences are mainly distributed in heterochromatic regions including the centromere. It was noticed that a few rDNA related repetitive sequences were found in this chromosome.
In addition to the cytological polymorphism revealed in this study, it is important to characterize the molecular organization of rDNAs in rice genomes. Since the nucleotide sequences of rDNAs and their spacers of several cultivated and wild species of rice have been registered in public databases, it is possible to survey more details of the variations in rice rDNA units. There were approximately 850 copies of 8-kb rDNA repeat unit per diploid genome of O. sativa ssp. japonica (Oono and Sugiura 1980). A detailed sequence analysis revealed that the telomeric region of the short arm of rice chromosome 9, the NOR site of japonica cv Nipponbare, consists of telomeric repeats, an rDNA array, and a retrotransposon-rich chromosomal region. Furthermore, more retrotransposons were found in the proximal rDNA-flanking region than in other subtelomeric regions or sequenced regions of the genome (Fujisawa et al. 2006). The ongoing wild species of rice genomic sequencing project (Oryza Map Alignment Project, OMAP, Wing et al. 2005) may soon provide enough information to resolve the mystery of the CCDD genome and the origins of rDNA polymorphism.
Rice chromosomes are small in size and lack accessible landmarks for identification of individual chromosome. A set of BAC clones has been evaluated to facilitate rice chromosome identification (Cheng et al. 2001a), however, those markers show less utility in labeling chromosomes of rice other than Nipponbare (O. sativa ssp. Japonica, personal experience). As mentioned above, high similarity in karyotypes were found among Oryza species (Chen and Wu 1982), we use the karyotype of IR36 (O. sativa ssp. indica) as a standard to identify individual chromosome of Oryza species in this study. However, the polymorphism in rDNA is insufficient for identifying chromosome complement of rice genomes. Except rDNAs, more molecular analyses together with cytological methods are needed to further understand the cyto-evolution in rice genomes.
References
Aggarwal R-K, Brar D-S, Nandi S, Huang N, Khush G-S (1999) Phylogenetic relationships among Oryza species revealed by AFLP markers. Theor Appl Genet 98:1320–1328
Cerbah M, Souza-Chies T, Jubier M-F, Lejeune B, Siljak-Yakovlev S (1998) Molecular phylogeny of the genus Hypochaeris using internal transcribed spacers of nuclear rDNA: inference for chromosomal evolution. Mol Biol Evol 15:345–354
Chen Y-C, Wu H-K (1982) A comparison of karyotypes among six Oryza species. Bot Bull Acad Sin 23:163–183
Cheng Z-K, Buell R, Wing R-A, Gu M-H, Jiang J (2001a) Toward a cytological characterization of the rice genome. Genome Res 11:2133–2141
Cheng Z, Presting G-G, Buell C-R, Wing R-A, Jiang J (2001b) High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157:1749–1757
Chung M-C, Ning C-N, Wu H-K (1993) Localization of ribosomal RNA genes on rice chromosomes. Bot Bull Acad Sin 34:43–55
Chung M-C, Wu H-K (1987) Karyotype analysis of IR36 and two trisomic lines of rice. Bot Bull Acad Sin 28:289–304
Comai L (2000) Genetic and epigenetic interactions in allopolyploid plants. Plant Mol Biol 43:387–399
Comai L, Tyagi A-P, Lysak M-A (2003) FISH analysis of meiosis in Arabidopsis allopolyploids. Chromosome Res 11:217–226
Cordesse F, Cooke R, Tremousaygue D, Grellet F, Delseny M (1993) Fine structure and evolution of the rDNA intergenic spacer in rice and other cereals. J Mol Evol 36:369–379
Dong F, Miller J-T, Jackson S-A, Wang G-L, Ronald P-C, Jiang J (1998) Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci USA 95:8135–8140
Feng Q, Zhang Y, Hao P, et al., (2002) Sequence and analysis of rice chromosome 4. Nature 420:316–320
Fujisawa M, Yamagata H, Kamiya K, Nakamura M, Saji S, Kanamori H, Wu J, Matsumoto T, Sasaki T (2006) Sequence comparison of distal and proximal ribosomal DNA arrays in rice (Oryza sativa L.) chromosome 9S and analysis of their flanking regions. Theor Appl Genet 113:419–428
Fukui K, Iijima K (1991) Somatic chromosome map of rice by imaging methods. Theor Appl Genet 81:589–596
Fukui K, Ohmido N, Khush G-S (1994) Variability in rDNA loci in genus Oryza detected through fluorescence in situ hybridization. Theor Appl Genet 87:893–899
Ge S, Sang T, Lu B-R, Hong D-Y (1999) Phylogeny of rice genomes with emphasis on regions of allotetraploid species. Proc Natl Acad Sci USA 96:14400–14405
Gerlach W-L, Bedbrook J-R (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucl Acids Res 7:1869–1885
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Kao F-I, Cheng Y-Y, Chow T-Y, Chen H-H, Liu S-M, Cheng C-H, Chung M-C (2006) An integrated map of Oryza sativa L. chromosome 5. Theor Appl Genet 112:891–902
Kurata N, Omura T (1978) Karyotype analysis in rice. A new method for identifying all chromosome pairs. Jpn J Genet 53:251–255
Leitch I-J, Heslop-Harrison J-S (1992) Physical mapping of the 18S-5.8-26S rRNA genes in barley by in situ hybridization. Genome 35:1013–1018
Li D, Zhang X (2002) Physical localization of the 18S-5.8S-26S rDNA and sequence analysis of ITS regions in Thinopyrum ponticum (Poaceae: Triticeae): implications for concerted evolution. Ann Bot 90:445–452
Linde-Laursen I, Ibsen E, Von Bothmer R, Giese H (1992) Physical localization of active and inactive rRNA gene loci in Hordeum marinum ssp. gussoneanum (4×) by in situ hybridization. Genome 35:1032–1036
Murata M, Shibata F, Yokota E (2006) The origin, meiotic behavior, and transmission of a novel minichromosome in Arabidopsis thaliana. Chromosoma 115:311–319
Ohmido N, Fukui K (1995) Cytological studies of African cultivated rice, Oryza glaberrima. Theor Appl Genet 91:212–217
Oono K, Sugiura M (1980) Heterogeneity of the ribosomal RNA gene clusters in rice. Chromosoma 76:85–89
Orgaard M, Heslop-Harrison J-S (1994) Relationships between species of Leymus, Psathyrostachys, and Hordeum (Poaceae, Triticeae) inferred from Southern hybridization of genomic and cloned DNA probes. Plant Sys Evol 189:217–231
Raina S-N, Mukai Y (1999) Detection of a variable number of 18S-5.8S-26S and 5S ribosomal DNA loci by fluorescent in situ hybridization in diploid and tetraploid Arachis species. Genome 42:52–59
Ricroch A, Peffley E-B, Baker R-J (1992) Chromosomal location of rDNA in Allium: in situ hybridization using biotin- and fluorescein-labeled probe. Theor Appl Genet 83:413–418
Sang Y, Liang G-H (2000) Comparative physical mapping of the 18S-5.8S-26S rDNA in three Sorghum species. Genome 43:918–922
Sano Y, Sano R (1990) Variation of the intergenic spacer regions of ribosomal DNA in cultivated and wild rice species. Genome 33:209–218
Schubert I, Wobus U (1985) In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92:143–148
Shishido R, Sano Y, Fukui K (2000) Ribosomal DNAs an exception to the conservation of gene order in rice genomes. Mol Gen Genet 263:586–591
Vaughan D-A, Morishima H, Kadowaki K (2003) Diversity in the Oryza genus. Curr Opin Plant Biol 6:139–146
Wang X, Shi X, Hao B, Ge S, Luo J (2005) Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol 165:937–946
Weider L-J, Elser J-J, Crease T-J, Mateos M, Cotner J-B, Markow T-A (2005) The functional significance of ribosomal (r)DNA variation: impacts on the evolutionary ecology of organisms. Annu Rev Ecol Evol Syst 36:219–242
Wing R-D, Ammiraju J-S-S, Luo M, Kim H, Yu Y, Kudrna D, Goicoechea J-L, Wang W, Nelson W, Rao K, Brar D, Mackill DJ, Han B, Soderlund C, Stein L, SanMiguel P, Jackson S (2005) The Oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol Biol 59:53–62
Yu J, Wang J, Lin W, Li SG, Li H, et al. (2005) The genomes of Oryza sativa: a history of duplications. PloS Biol 3:266–281
Acknowledgments
The authors are thankful to Dr. Jiming Jiang (Department of Horticulture, University of Wisconsin–Madison, USA) for providing plasmids pTA71 and pRCS2. This work was supported by grants from the National Science Council of Taiwan (NSC-95-2313-B-001-019) and from Institute of Plant and Microbial Biology, Academia Sinica, Taiwan to Mei-Chu Chung.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.S. Heslop-Harrison.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Chung, MC., Lee, YI., Cheng, YY. et al. Chromosomal polymorphism of ribosomal genes in the genus Oryza . Theor Appl Genet 116, 745–753 (2008). https://doi.org/10.1007/s00122-007-0705-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0705-z