Abstract
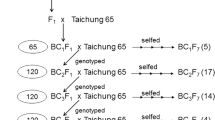
Rice (Oryza sativa L.) chromosome segment substitution lines (CSSLs), in which chromosomal segments of the Indian landrace “Kasalath” replace the corresponding endogenous segments in the genome of the Japanese premium rice “Koshihikari”, are available and together cover the entire genome. Chromosome regions affecting a trait (CRATs) can be identified by comparison of phenotypes with genotypes of CSSLs. We detected 99 CRATs for 15 agronomic or morphological traits. “Kasalath” had positively acting alleles in 53 CRATs. Its CRATs increased panicle number per plant by up to 23.3%, grain number per panicle by up to 30.8%, and total grain number by up to 15.1%, relative to “Koshihikari”. CRATs were identified for grain size (grain thickness and width), with positive effects of about 5.0%. A CRAT on chromosome 8 almost doubled the weight of roots in uppermost soil layers compared to “Koshihikari”. Additionally, “Kasalath” possessed CRATs for higher lodging resistance (reduction in plant height and increase in stem diameter). In some cases, multiple CRATs were detected in the same chromosome regions. Therefore, CSSLs with these chromosome segments might be useful breeding materials for the simultaneous improvement of multiple traits. Five CRATs, one for plant height on chromosome 1, one for stem diameter on chromosome 8, and three for heading date on chromosomes 6, 7, and 8 overlapped with the corresponding QTLs that already had been mapped with back-crossed inbred lines of “Nipponbare” and “Kasalath”. In both “Koshihikari” CRATs and “Nipponbare” QTLs, “Kasalath” had similar effects.



Similar content being viewed by others
References
Ashikari M, Matsuoka M (2006) Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Trend Plant Sci 11:344–350
Debi BR, Mushika J, Taketa S, Miyao A, Hirochika H, Ichii M (2003) Isolation and characterization of a short lateral root mutant in rice (Oryza sativa L.). Plant Sci 165:895–903
Ebitani T, Takeuchi Y, Nonoue Y, Yamamoto T, Takeuchi K, Yano M (2005) Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segment of indica rice cultivar “Kasalath” in a genetic background of japonica elite cultivar “Koshihikari”. Breed Sci 55:65–73
Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162
Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin SY, Antonio BA, Parco A, Kajiya H, Huang N, Yamamoto K, Nagamura Y, Kurata N, Khush GS, Sasaki T (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148:479–494
Hittalmani S, Huang N, Courtois B, Venuprasad R, Shashidhar HE, Zhuang J-Y, Zheng K-L, Liu G-F, Wang G-C, Sidhu JS, Srivantaneeyakul S, Singh VP, Bagali PG, Prasanna HC, McLaren G, Khush GS (2003) Identification of QTL for growth- and grain yield-related traits in rice across nine locations of Asia. Theor Appl Genet 107:679–690
Howell PM, Marshall DF, Lydiate DJ (1996) Towards developing intervarietal substitution lines in Brassica napus using marker-assisted selection. Genome 39:348–358
Ishikawa S, Ae N, Yano M (2005a) Chromosomal regions with quantitative trait loci controlling cadmium concentration in brown rice (Oryza sativa). New Phytol 168:345–350
Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005b) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46:79–86
Ishimaru K (2003) Identification of a locus increasing rice yield and physiological analysis of its function. Plant Physiol 133:1083–1090
Ishimaru K, Yano M, Aoki N, Ono K, Hirose T, Lin SY, Monna L, Sasaki T, Ohsugi R (2001) Toward the mapping of physiological and agronomic characters on a rice function map: QTL analysis and comparison between QTLs and expressed sequence tags. Theor Appl Genet 102:793–800
Ishimaru K, Ono K, Kashiwagi T (2004) Identification of a new gene controlling plant height in rice using the candidate-gene strategy. Planta 218:388–395
Ishimaru K, Kashiwagi T, Hirotsu N, Madoka Y (2005) Identification and physiological analyses of a locus for rice yield potential across the genetic background. J Exp Bot 56:2745–2753
Kashiwagi T, Ishimaru K (2004) Identification and functional analysis of a locus for improvement of lodging resistance in rice. Plant Physiol 134:676–683
Kashiwagi T, Hirotsu N, Madoka Y, Ookawa T, Ishimaru K (2007) Improvement of resistance to bending-type lodging in rice. Jpn J Crop Sci 76:1–9
Katagiri S, Wu J, Ito Y, Karasawa W, Shibata M, Kanamori H, Katayose Y, Namiki N, Matsumoto T, Sasaki T (2004) End sequencing and chromosomal in silico mapping of BAC clones derived from an indica rice cultivar, Kasalath. Breed Sci 54:273–279
Kojima Y, Ebana K, Fukuoka S, Nagamine T, Kawase M (2005) Development of an RFLP-based rice diversity research set of germplasm. Breed Sci 55:431–440
Kondo M, Pablico PP, Aragones DV, Agbisit R, Abe J, Morita S, Courtois B (2003) Genotypic and environmental variations in root morphology in rice genotypes under upland field condition. Plant Soil 255:189–200
Koumproglou R, Wilkes TM, Townson P, Wang XY, Beyson J, Pooni HS, Newbury HJ, Kearsey MJ (2002) STAIRS: a new genetic resource for functional genomic studies of Arabidopsis. Plant J 31:355–364
Lin HX, Yamamoto T, Sasaki T, Yano M (2000) Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines. Theor Appl Genet 101:1021–1028
Lin HX, Liang ZW, Sasaki T, Yano M (2003) Fine mapping and characterization of a quantitative trait locus, Hd4 and Hd5, controlling heading date in rice. Breed Sci 53:51–59
Matus I, Corey A, Filichkin T, Hayes PM, Vales MI, Kling J, Riera-Lizarazu O, Sato K, Powell W, Waugh R (2003) Development and characterization of recombinant chromosome substitution lines (RCSLs) using Hordeum vulgare subsp. spontaneum as a source of donor alleles in a Hordeum vulgare subsp. vulgare background. Genome 46:1010–1023
McKendry AL, Tague DN, Finney PL, Miskin KE (1996) Effect of 1BL.1RS on milling and baking quality of soft red winter wheat. Crop Sci 36:848–851
Mei HW, Xu JL, Li ZK, Yu XQ, Guo LB, Wang YP, Ying CS, Luo LJ (2006) QTLs influencing panicle size detected in two reciprocal introgressive line (IL) populations in rice (Oryza sativa L.). Theor Appl Genet 112:648–656
Morita S, Yamada S, Abe J (1995) Analysis on root system morphology in rice with reference to varietal differences at ripening stage. Jpn J Crop Sci 64:58–65
Servin B, Martin OC, Mézard M, Hospital F (2004) Toward a theory of marker-assisted gene pyramiding. Genetics 168:513–523
Tanaka N, Kubota F, Abiru H (1985) A new core-sampling method in examining rice root system in paddy fields. Jpn J Crop Sci 54:379–386
Tanksley SD (1993) Mapping polygenes. Ann Rev Genet 27:205–233
Tian F, Li DJ, Fu Q, Zhu ZF, Fu YC, Wang XK, Sun CQ (2006) Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor Appl Genet 112:570–580
Toojinda T, Baird E, Booth A, Broers L, Hayes P, Powell W, Thomas W, Vivar H, Yong G (1998) Introgression of quantitative trait loci (QTLs) determining stripe rust resistance in barley: an example of marker-assisted line development. Theor Appl Genet 96:123–131
Tripathi SC, Sayre KD, Kaul JN, Narang RS (2003) Growth and morphology of spring wheat (Triticum aestivum L.) culms and their association with lodging: effects of genotypes, N levels and ethephon. Field Crops Res 84:271–290
Uchiyamada H (1995) Breeding in rice cv. Koshihikari. Koshihikari. In: Oritani T et al (eds) Koshihikari. Nobunkyo, Tokyo, pp 66–72
Won JG, Hirahara Y, Yoshida T, Imabayashi S (1998) Selection of rice lines using SPGP seedling method for direct seeding. Plant Prod Sci 1:280–285
Xiao J, Grandillo S, Ahn SN, McCouch SR, Tanksley SD, Li J, Yuan L (1996) Genes from wild rice improve yield. Nature 384:223–224
Yano M, Harushima Y, Nagamura Y, Kurata N, Minobe Y, Sasaki T (1997) Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theor Appl Genet 95:1025–1032
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2743–2483
Zhang X, Zhou S, Fu Y, Su Z, Wang X, Sun C (2006) Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol Biol 62:247–259
Zuber U, Winzeler H, Messmer MM, Keller B, Schmid JE, Stamp P (1999) Morphological traits associated with lodging resistance of spring wheat (Triticum aestivum L.). J Agron Crop Sci 182:17–24
Acknowledgments
This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN). The authors acknowledge the kind gift of CSSL seeds by the Rice Genome Resource Center in Japan and the National Institution of Agrobiological Sciences, and Dr. Kiyoshi Nagasuga (National Agricultural Research Center for Tohoku) for filed works.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Xue.
Both Y. Madoka and T. Kashiwagi have contributed equally to this article.
Rights and permissions
About this article
Cite this article
Madoka, Y., Kashiwagi, T., Hirotsu, N. et al. Indian rice “Kasalath” contains genes that improve traits of Japanese premium rice “Koshihikari”. Theor Appl Genet 116, 603–612 (2008). https://doi.org/10.1007/s00122-007-0693-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0693-z