Abstract
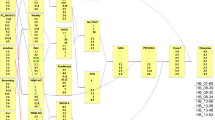
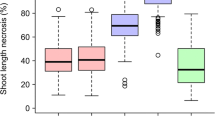
Although fire blight, caused by the bacterium Erwinia amylovora, is one of the most destructive diseases of apple (Malus × domestica) worldwide, no major, qualitative gene for resistance to this disease has been identified to date in apple. We conducted a quantitative trait locus (QTL) analysis in two F1 progenies derived from crosses between the cultivars Fiesta and either Discovery or Prima. Both progenies were inoculated in the greenhouse with the same strain of E. amylovora, and the length of necrosis was scored 7 days and 14 days after inoculation. Additive QTLs were identified using the mapqtl software, and digenic epistatic interactions, which are an indication of putative epistatic QTLs, were detected by two-way analyses of variance. A major QTL explaining 34.3–46.6% of the phenotypic variation was identified on linkage group (LG) 7 of Fiesta in both progenies at the same genetic position. Four minor QTLs were also identified on LGs 3, 12 and 13. In addition, several significant digenic interactions were identified in both progenies. These results confirm the complex polygenic nature of resistance to fire blight in the progenies studied and also reveal the existence of a major QTL on LG7 that is stable in two distinct genetic backgrounds. This QTL could be a valuable target in marker-assisted selection to obtain new, fire blight-resistant apple cultivars and forms a starting point for discovering the function of the genes underlying such QTLs involved in fire blight control.


Similar content being viewed by others
References
Ahmadi N, Albar L, Pressoir G, Pinel A, Fargette D, Ghesquiere A (2001) Genetic basis and mapping of the resistance to rice yellow mottle virus. III. Analysis of QTL efficiency in introgressed progenies confirmed the hypothesis of complementary epistasis between two resistance QTL. Theor Appl Genet 103:1084–1092
Alston FH, Phillips KL, Evans KM (2000) A Malus gene list. Acta Hortic 538:561–570
Baldi P, Patocchi A, Zini E, Toller C, Velasco R, Komjanc M (2004) Cloning and mapping of resistance gene homologues in apple. Theor Appl Genet 109:231–239
Bonn WG, Van der Zwet T (2000) Distribution and economic importance of fire blight. In: Vanneste J-L (ed) Fire blight: the disease and its causative agent: Erwinia amylovora. CAB Int, Wallingford, pp 37–53
Calenge F, Faure A, Drouet D, Parisi L, Brisset M-N, Paulin J-P, Van der Linden CG, Van de Weg WE, Schouten H, Lespinasse Y, Durel C-E (2004a) Genomic organization of resistance factors against scab (Venturia inaqualis), powdery mildew (Podosphaera leucotricha) and fire blight (Erwinia amylovora) in apple. In: Tikhonovitch I, Lugtenberg B, Provorov N (eds) Proc 11th Int Congr Mol Plant-Microbe Interact. Biol Plant Microbe Interact 4:35–39
Calenge F, Faure A, Goerre M, Gebhardt C, Van de Weg WE, Parisi L, Durel C-E (2004b) A QTL analysis reveals both broad-spectrum and isolate-specific QTL for scab resistance in an apple progeny challenged with eight isolates of Venturia inaequalis. Phytopathology 94:370–379
Calenge F, Van der Linden CG, Van de Weg WE, Schouten HJ, Van Arkel G, Denancé C, Durel C-E (2005) Resistance gene analogues identified through the NBS-profiling method map close to major genes and QTL for disease resistance in apple. Theor Appl Genet 110:660–668
Conner PJ, Brown SK, Weeden NF (1997) Randomly amplified polymorphic DNA-based genetic linkage maps of three apple cultivars. J Am Soc Hortic Sci 3:350–359
Dondini L, Pierantoni L, Gaiotti F, Chiodini R, Tartarini S, Bazzi C, Sansavini S (2004) Identifying QTLs for fire blight resistance via a European pear (Pyrus communis L.) genetic linkage map. Mol Breed 14:407–418
Durel C-E, Parisi L, Laurens F, Van de Weg E, Liebhard R, Koller B, Jourjon MF (2003) Genetic dissection of partial resistance against two monoconidial strains of the new race 6 of Venturia inaequalis in apple. Genome 46:224–234
Gao ZS, Van de Weg WE, Schaart JG, Schouten HJ, Tran DH, Kodde L, Van der Meer IM, Van der Geest AHM, Kodde J, Breiteneder H, Hoffmann-Sommergruber K, Bosch D, Gilissen LJWJ (2005) Genomic cloning and linkage mapping of the Mal d 1 (PR-10) gene family in apple (Malus × domestica) Theor Appl Genet (in press)
Gebhardt C, Valkonen JPT (2001) Organization of genes controlling disease resistance in the potato genome. Annu Rev Phytopathol 39:79–102
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Hemmat M, Brown SK (2002) Tagging and mapping scab resistance genes from R12740-7A Apple. J Am Soc Hortic Sci 127:365–370
Hemmat M, Weeden NF, Manganaris AG, Lawson DM (1994) Molecular linkage map for apple. J Hered 85:4–11
Jones AL, Schnabel E (2000) The development of streptomycin-resistant strains of Erwinia amylovora. In: Vanneste J-L (ed) Fire blight: the disease and its causative agent: Erwinia amylovora. CAB Int, Wallingford, pp 335–367
King GJ, Alston FH, Batlle I, Chevreau E, Gessler C, Janse J, Lindhout P, Manganaris AG, Sansavini S, Schmidt H, Tobutt KR (1991) “The European Apple Genome mapping Project”—developing a strategy for mapping genes coding for agronomic characters in tree species. Euphytica 56:89–94
Korban SS, Janick J, Williams EB, Emerson FH (1988) “Dayton” apple. HortScience 23:927–928
Le Lezec M, Thibault B, Balavoine P, Paulin J-P (1985) Sensibilité variétale du pommier et du poirier. Phytoma Février:37–44
Le Lezec M, Babin J, Lecomte P (1990) Le feu bactérien: sensibilité de nouvelles variétés de pommes, premiers essais. L’arboriculture Fruitière 434:24–32
Liebhard R, Koller B, Gianfranceschi L, Gessler C (2003a) Creating a saturated reference map for the apple (Malus × domestica Borkh.) genome. Theor Appl Genet 106:1497–1508
Liebhard R, Koller B, Kellerhalls M, Pfammatter W, Jermini M, Gessler C (2003b) Mapping quantitative field resistance in apple against scab. Phythopathology 93:493–501
Lespinasse Y, Aldwinckle H (2000) Breeding for resistance to fire blight. In: Vanneste J-L (ed) Fire blight: the disease and its causative agent: Erwinia amylovora. CAB Int, Wallingford, pp 253–273
Lespinasse Y, Durel C-E, Laurens F, Parisi L, Chevalier M, Pinet C (2000) A European project: DARE—durable apple resistance in Europe (FAIR5 CT97-3898) durable resistance of apple to scab and powdery-mildew: one step more towards an environmental friendly orchard. Acta Hortic 538:197–200
Maliepaard C, Alston F, Van Arkel G, Brown L, Chevreau E, Dunemann F, Evans KM, Gardiner S, Guilford P, Van Heusden AW, Janse J, Laurens F, Lynn JR, Manganaris AG, Den Nijs APM, Periam N, Rikkerink E, Roche P, Ryder C, Sansavini S, Schmidt H, Tartarini S, Verhaegh JJ, Vrielink-Van Ginkel M, King GJ (1998) Aligning male and female linkage maps of apple (Malus pumila) using multi-allelic markers. Theor Appl Genet 97:60–73
Manzanares-Dauleux MJ, Delourme R, Baron F, Thomas G (2000) Mapping of one major gene and of QTLs involved in resistance to clubroot in Brassica napus. Theor Appl Genet 101:885–891
McDowell JM, Woffenden BJ (2003) Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol 21:178–183
McMullen M, Simcox KD (1995) Genomic organization of disease and insect resistance genes in Maize. Mol Plant Microbe Interact 8:811–815
Miklas PN, Delorme R, Stone V, Daly MJ, Stavely JR, Steadman JR, Bassett MJ, Beaver JS (2000) Bacterial, fungal, and viral disease resistance loci mapped in a recombinant inbred common bean population (“Dorado”/XAN 176). J Am Soc Hortic Sci 125:476–481
Norelli JL, Aldwinckle HS, Beer SV (1984) Differential host × pathogen interactions among cultivars of apple and strains of Erwinia amylovora. Phytopathology 74:136–139
Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, Zamir D, Fluhr R (1997) The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine rich repeat superfamily of plant resistance genes. Plant Cell 9:521–532
Phipps JB, Robertson KR, Rohrer JR (1991) Origins and evolution of subfamily Maloideae (Rosaceae). Syst Bot 16:303–332
Thomson S (2000) Epidemiology of fire blight. In: Vanneste J-L (ed) Fire blight: the disease and its causative agent: Erwinia amylovora. CAB Int, Wallingford, pp 370–382
Vanneste J-L (2000) What is fire blight? Who is Erwinia amylovora? How to control it? In: Vanneste J-L (ed) Fire blight: the disease and its causative agent: Erwinia amylovora. CAB Int, Wallingford, pp 1–6
Van Ooijen JW, Maliepaard C (1996) mapqtl version 3.0: software for the calculation of QTL positions on genetic maps. CPRO-DLO, Wageningen
Venisse JS, Malnoy M, Faize M, Paulin JP, Brisset MN (2002) Modulation of defence responses of Malus spp during compatible and incompatible interactions with Erwinia amylovora. Mol Plant Microbe Interact 15:1204–1212
Wang GL, Ruan DL, Song WY, Sideris S, Chen LL, Pi LY, Zhang S, Zhang Z, Fauquet C, Gaut B, Ronald P (1998) Xa21D encodes a receptor-like molecule with a leucine-rich-repeat domains that determine race specific recognition and is subject to adaptative evolution. Plant Cell 10:765–779
Yamamoto T, Kimura T, Saito T, Kotobuki K, Matsuta N, Liebhard R, Gessler C, Van de Weg WE, Hayashi T (2004) Genetic linkage maps of Japanese and European pears aligned to the apple consensus map. Acta Hortic 663:51–56
Young N (1996) QTL mapping and quantitative disease resistance in plants. Annu Rev Phytopathol 34:479–501
Acknowledgements
We thank Roland Chartier and Lysiane Leclout for their excellent technical work in the greenhouse. This research was based on unpublished results from the European project FAIR5 CT97-3898 (DARE: Durable Apple Resistance in Europe). It was partly supported by a grant from INRA/Région Pays de la Loire, France, allocated to the first author.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Nybom
Rights and permissions
About this article
Cite this article
Calenge, F., Drouet, D., Denancé, C. et al. Identification of a major QTL together with several minor additive or epistatic QTLs for resistance to fire blight in apple in two related progenies. Theor Appl Genet 111, 128–135 (2005). https://doi.org/10.1007/s00122-005-2002-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-2002-z