Abstract
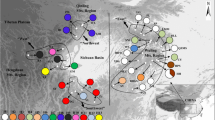
Sequences of chloroplast DNA (cpDNA) atpB–rbcL intergenic spacers of individuals of a tree fern species, Alsophila spinulosa, collected from ten relict populations distributed in the Hainan and Guangdong provinces, and the Guangxi Zhuang region in southern China, were determined. Sequence length varied from 724 bp to 731 bp, showing length polymorphism, and base composition was with high A+T content between 63.17% and 63.95%. Sequences were neutral in terms of evolution (Tajima’s criterion D=−1.01899, P>0.10 and Fu and Li’s test D*=−1.39008, P>0.10; F*=−1.49775, P>0.10). A total of 19 haplotypes were identified based on nucleotide variation. High levels of haplotype diversity (h=0.744) and nucleotide diversity (Dij=0.01130) were detected in A. spinulosa, probably associated with its long evolutionary history, which has allowed the accumulation of genetic variation within lineages. Both the minimum spanning network and neighbor-joining trees generated for haplotypes demonstrated that current populations of A. spinulosa existing in Hainan, Guangdong, and Guangxi were subdivided into two geographical groups. An analysis of molecular variance indicated that most of the genetic variation (93.49%, P<0.001) was partitioned among regions. Wright’s isolation by distance model was not supported across extant populations. Reduced gene flow by the Qiongzhou Strait and inbreeding may result in the geographical subdivision between the Hainan and Guangdong + Guangxi populations (FST=0.95, Nm=0.03). Within each region, the star-like pattern of phylogeography of haplotypes implied a population expansion process during evolutionary history. Gene genealogies together with coalescent theory provided significant information for uncovering phylogeography of A. spinulosa.



Similar content being viewed by others
References
Beebee T, Rowe G (2004) An introduction to molecular ecology. Oxford University Press, New York
Bennett KD (1990) Milankovitch cycles and their effects on species in ecological and evolutionary time. Paleobiology 16:11–21
Brown AHD (1979) Enzyme polymorphism in plant populations. Theor Popul Biol 15:1–42
Castelloe J, Templeton AR (1994) Root probability for intraspecific gene trees under neutral coalescent theory. Mol Phylogenet Evol 3:102–113
Cheng ZY, Tao GD, Xu ZF (1990) A study on the biological characteristics and the endangering factors of Alsophila spinulosa. Acta Bot Yunnanica 12:186–190
Chiang TY, Schaal BA (1999) Phylogeography of ten North American Hylocomium splendens based on nrDNA ITS sequences. Mol Ecol 8:1037–1042
Chiang TY, Schaal BA, Peng CI (1998) Universal primers for amplification and sequencing a noncoding spacer between the atpB and rbcL genes of chloroplast DNA. Bot Bull Acad Sin 39:245–250
Chiang TY, Chiang YC, Chen YJ, Chou CH, Havanond S, Hong TN, Huang S (2001) Phylogeography of Kandelia candel in east Asiatic mangroves based on nucleotide variation of chloroplast and mitochondrial DNAs. Mol Ecol 10:2697–2710
Collinson ME (1996) In: Camus JM, Gibby M, Johns RJ (eds) Pteridology in perspective. Royal Botanic Garden, Kew, pp 349–394
Conant DS (1990) Observations on the reproductive biology of Alsophila species and hybrids (Cyatheaceae). Ann Mo Bot Gard 77:290–296
Demesure B, Comps B, Petit RJ (1996) Chloroplast DNA phylogeography of the common beech (Fagus sylvatica L.) in Europe. Evolution 50:2515–2520
Dynesius M, Jansson R (2000) Evolutionary consequences of changes in species’ geographical distribution driven by Milankovitch climate oscillations. Proc Natl Acad Sci USA 97:9115–9120
Eriksson T (2004) Ferns reawakened. Nature 428:480–481
Excoffier L, Smouse PE (1994) Using allele frequencies and geographic subdivision to reconstruct gene trees with a species: molecular variance parsimony. Genetics 136:343–359
Felsenstein J (1995) PHYLIP (Phylogeny Inference Package) version 3.57. University of Washington, Seattle
Ferris C, King RA, Vainola R, Hewitt GM (1998) Chloroplast DNA recognizes three refugia sources of European oaks and suggests independent eastern and western immigrations to Finland. Heredity 80:584–593
Fu LG (1991) The Red List of Chinese plants. Science, Beijing
Gottlieb LD (1981) Electrophoretic evidence and plant populations. Prog Phytochem 7:1–45
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hamrick JL, Linhart YB, Mitton JB (1979) Relationships between life history characteristics and electrophoretically detectable genetic variation in plants. Ann Rev Ecol Syst 10:173–200
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Huang S, Chiang YC, Schaal BA, Chou CH, Chiang TY (2001) Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Mol Ecol 10:2669–2681
Hudson RR, Slatkin M, Maddison WP (1992) Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589
Hwang SY, Lin TP, Ma CS, Lin CL, Chung JD, Yang JC (2003) Postglacial population growth of Cunninghamia konishii (Cupressaceae) inferred from phylogeographical and mismatch analysis of chloroplast DNA variation. Mol Ecol 12:2689–2695
Kimura M, Weiss GH (1964) The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics 49:561–576
Larena BG, Aguilar JF, Feliner GN (2002) Glacial-induced altitudinal migrations in Armeria (Plumbaginaceae) inferred from patterns of chloroplast DNA haplotype sharing. Mol Ecol 12:1965–1974
Li WH (1997) Molecular evolution. Sinauer, Sunderland, pp 402–408
Li JW and Haufler CH (1999) Genetic variation, breeding systems, and patterns of diversification in Hawaiian Polypodium (Polypodiaceae). Syst Bot 24:339–355
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Ann Rev Ecol Syst 15:65–95
Lu SY, Peng CI, Cheng YP, Hong KH, Chiang TY (2001) Chloroplast DNA phylogeography of Cunninghamia konishii (Cupressaceae) an endemic conifer of Taiwan. Genome 44:767–807
Lu SY, Hong KH, Liu SL, Cheng YP, Wu WL, Chiang TY (2002) Genetic variation and population differentiation of Michelia formosana (Magnoliaceae) based on cpDNA variation and RAPD fingerprints: relevance to post-Pleistocene recolonization. J Plant Res 115:203–216
Maki M, Asada Y (1988) High genetic variability revealed by allozymic loci in the narrow endemic fern Polystichum otomasui (Dryopteridaceae). Heredity 80:604–610
Maruyama T (1971) Analysis of population structure. II. Two-dimensional stepping stone models of finite length and other geographically structured populations. Ann Hum Genet 35:179–196
Nagylaki T (1976) The decay of genetic variability in geographically structured populations. II. Theor Popul Biol 10:70–82
Pages RDM, Holmes E (1998) Molecular evolution. A phylogenetic approach. Blackwell, Oxford, pp 201–227
Petit RJ, Pineau E, Demesure B, Bacilieri R, Ducousso A, Kremer A (1997) Chloroplast DNA footprints of postglacial recolonization by oaks. Proc Natl Acad Sci USA 94:9996–10001
Pryor KV, Young JE, Rumsey FJ, Edwards KJ, Bruford MW, Rogers HJ (2001) Diversity, genetic structure and evidence of outcrossing in British populations of the rock fern Adiantum capillus-veneris using microsatellites. Mol Ecol 10:1881–1894
Ranker TA, Gemmill CEC, Trapp PG (2000) Microevolutionary patterns and processes of the native Hawaiian colonizing fern Odontosoria chinensis (Lindsaesceae). Evolution 54:829–839
Rothwell GW (1996) In: Camus JM, Gibby M, Johns RJ (eds) Pteridology in perspective. Royal Botanic Garden, Kew, pp 395–404
Rozas J, Rozas R (1999) DNASP version 3.0: an integrated program for molecular population genetics and molecular evolution genetic analysis. Bioinformatics 15:174–175
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Mol Ecol 7:465–474
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva
Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallon S, Lupia R (2004) Ferns diversified in the shadow of angiosperms. Nature 428:553–557
Schneller J, Holderegger R (1996) Colonization events and genetic variability within populations of Asplenium ruta-muraria. In: Camus JM, Gibby M, Johns RJ (eds) Pteridology in perspective. Royal Botanic Gardens, London, pp 571–580
Skog JE (2001) Biogeography of mesozoic leptosporangiate ferns related to extant ferns. Brittonia 53:236–269
Slatkin M (1993) Isolation by distance in equilibrium and nonequilibrium populations. Evolution 47:264–279
Smith AR (1972) Comparison of fern and flowering plant distribution with some evolutionary interpretations for ferns. Biotropica 4:4–9
Soltis PS, Soltis DE (1988) Genetic variation and population genetic structure in the fern Blechnum spicant (Blechnaceae) from western North America. Am J Bot 75:37–44
Su YJ, Wang T, Yang WD, Huang C, Fan GK (1998) DNA extraction and RAPD analysis of Podocarpus. Acta Sci Nat Univ Sunyatseni 37:13–18
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Trewick SA, Morgan-Richards M, Russell SJ, Henderson S, Rumsey FJ, Pinter I, Barrett JA, Gibby M, Vogel JC (2002) Polyploidy, phylogeography and Pleistocene refugia of the rockfern Asplenium ceterach: evidence from chloroplast DNA. Mol Ecol 11:2003–2012
Tryon RM (1970) The classification of Cyatheaceae. Contrib Gray Herb 200:3–53
Tryon AF, Lugardon B (1991) Spores of the Pteridophyta: surface, wall structure, and diversity based on electron microscope studies. Springer, Berlin Heidelberg New York
Tryon RM, Tryon AF (1982) Ferns and allied plants. Springer, Berlin Heidelberg New York
Vogel JC, Rumsey FJ, Russell SJ, Cox CJ, Holmes JS, Bujnoch W, Stark C, Barrett JA, Gibby M (1999a) Genetic structure, reproductive biology and ecology of isolated populations of Asplenium csikii (Aspleniaceae, Pteridophyta). Heredity 83:604–612
Vogel JC, Rumsey FJ, Schneller JJ, Barrett JA, Gibby M (1999b) Where are the glacial refugia in Europe? Evidence from pteridophytes. Biol J Linn Soc 66:23–37
Wang JH, Huang YJ, Shi GL (1996) Reproduction and growth of Alsophila spinulosa in Dinghushan mountain. Guihaia 16:283–286
Wang JY, Yao SH, Cheng GZ (1997) Researching on the karyotype of Alsophila spinulosa (Hook) Tryon of Guizhou. J Guizhou Normal Univ (Nat Sci) 15:21–25
Wang T, Su YJ, Li XY, Zheng B, Wang BS (2003a) RAPD analysis of the genetic variation within populations of a relict tree fern, Alsophila spinulosa (Cyatheaceae). Acta Ecol Sin 23:160–165
Wang T, Su YJ, Zheng B, Chen GP, Zeng QL (2003b) Phylogenetic analysis of the chloroplast trnL intron and trnL-trnF intergenic spacer sequences of the Cyatheaceae plants from China. J Trop Subtrop Bot 11:137–142
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Wright S (1943) Isolation by distance. Genetics 28:139–156
Xing FW, Wu TL, Li ZX, Ye HG, Chen BH (1995) Endemic plants of Hainan island. J Trop Subtrop Bot 3:1–12
Zanten BO van (1978) Experimental studies of trans-oceanic long-range dispersal of moss spores in the Southern Hemisphere. J Hatton Bot Lab 44:455–482
Zoller S, Lutzoni F, Scheidegger C (1999) Genetic variation within and among populations of the threatened lichen Lobaria pulmonaria in Switzerland and implications for its conservation. Mol Ecol 8:2049–2059
Acknowledgments
We thank Dr. George Littlejohn at Plant Science Group, IBLS/Biochemistry and Molecular Biology, University of Glasgow and Ms. Elaine Goodman at Sun Yat-sen University for advice and revision of the manuscript. This research was supported by grants from National Natural Science Foundation of China (grant no. 30170101), the Key Project of National Natural Science Foundation of China (grant no. 39830310), and the Natural Science Foundation of Guangdong Province, China (grant no. 011125).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D.B. Neale
Rights and permissions
About this article
Cite this article
Su, Y., Wang, T., Zheng, B. et al. Population genetic structure and phylogeographical pattern of a relict tree fern, Alsophila spinulosa (Cyatheaceae), inferred from cpDNA atpB–rbcL intergenic spacers. Theor Appl Genet 109, 1459–1467 (2004). https://doi.org/10.1007/s00122-004-1761-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1761-2