Abstract
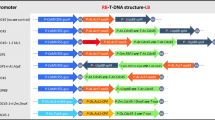
To investigate the potential of heterologous transposons as a gene-tagging system in broccoli (Brassica oleracea var. italica), we have introduced a Dissociation (Ds)-based two-element transposon system. Ds has been cloned into a 35S-SPT excision-marker system, with transposition being driven by an independent 35S-transposase gene construct. In three successive selfed generations of plants, there was no evidence of germinal-excision events. In a previous study, we overcame this apparent inability to produce B. oleracea plants with germinal excisions by performing a novel tissue-culture technique to select for fully green shoots from seed with somatic excision events. The results showed a very high efficiency of regeneration of fully green plants (up to 65%), and molecular analysis showed that the plants contained the equivalent of a germinal-excision event. In this study, we followed the previous work by using inverse and nested PCR to generate probes of flanking genomic DNA adjacent to independently reinserted Ds elements, and these were hybridised to DNA from a double-haploid mapping population of B. oleracea. Seventeen Ds insertions and the original Ds T-DNA site have been localised, and these are spread over six (out of nine) linkage groups. Distribution of inserts show that 15 were found on a different linkage group to the original ‘launch’ site, and of these 11 were found to be clustered on two separate groups. Previous studies in other plant species have found that germinal excision of Ds predominantly moves to sites linked close to the donor site. However, this study shows a potential to produce plants with Ds insertion scattered over many unlinked sites.






Similar content being viewed by others
References
Altmann T, Felix G, Jessop A, Kauschmann A, Uwer U, Pina-Cortés H, Willmitzer L (1995) Ac/Ds transposon mutagenesis in Arabidopsis thaliana: mutant spectrum and frequency of Ds insertion mutant. Mol Gen Genet 247:646–652
Baker B, Schell J, Lörz H, Federoff N (1988) Transposition of the maize controlling element Activator in tobacco. Proc Natl Acad Sci USA 83:4844–4848
Bancroft I, Dean C (1993) Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics 134:1221–1229
Bancroft I, Jones JDG, Dean C (1993) Heterologous transposon tagging of the DRL1 locus in Arabidopsis. Plant Cell 5:631–638
Belzile F, Yoder JI (1992) Pattern of somatic transposition in a high-copy Ac tomato line. Plant J 2:173–179
Bennett MD (1996) The nucleotype, the natural karyotype and the ancestral genome. Symp Soc Exp Biol 50:45–52
Biezen EA van der, Brandwagt BF, Van Leeuwen W, Nijkamp HJJ, Hille J (1996a) Identification and isolation of the FEEBLY gene from tomato by transposon tagging. Mol Gen Genet 251:267–280
Biezen EA van der, Cardol EF, Chung HY, Nijkamp HJJ, Hille J (1996b) Frequency and distance of transposition of a modified Dissociation element in transgenic tobacco. Transgenic Res 5:343–357
Bhatt AM, Page T, Lawson EJR, Lister C, Dean C (1996) Use of Ac as an insertional mutagen in Arabidopsis. Plant J 9:935–945
Bohuon EJR, Keith DJ, Parkin IAP, Sharpe AG, Lydiate DJ (1996) Alignment of the conserved C genome of Brassica oleracea and Brassica napus. Theor Appl Genet 93:833–839
Carroll BJ, Klimyuk VI, Thomas CM, Bishop GJ, Harrison K, Scofield SR, Jones JDG (1995) Germinal transposition of the maize element Dissociation from T-DNA loci in tomato. Genetics 139:407–420
Covey SN, Hull R (1981) Transcription of cauliflower mosaic virus DNA. Detection of transcripts, properties, and location of the gene encoding the virus inclusion body protein. Virology 111:463–474
De Block MD, De Brouwer D, Tenning P (1989) Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the expression of the Bar and neo genes in the transgenic plants. Plant Physiol 91:694–701
Dooner HK, Keller J, Harper E, Ralston E (1991) Variable patterns of transposition of the maize element Activator in tobacco. Plant Cell 3:473–482
Feinberg AP, Vogelstein B (1984) A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 137:266–267
Haring MA, Gao J, Volbeda T, Rommens CMT, Nijkamp HJJ, Hille J (1989) A comparative study of Tam 3 and Ac transposition in transgenic tobacco and petunia plants. Plant Mol Biol 13:189–201
Healy J, Corr C, De Young J, Baker B (1993) Linked and unlinked transposition of a genetically marked Dissociation element in transgenic tomato. Genetics 134:571–584
Izawa T, Miyazaki C, Yamamoto M, Terada R, Iida S, Shimamoto K (1991) Introduction and transposition of the maize transposable element Ac in rice (Oriza sativa L.). Mol Gen Genet 227:391–396
Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG (1994) Isolation of the tomato Cf-9 gene for resistance to Clodospurium fuluum by transposition tagging. Science 266:789–793
Jones JDG, Carland F, Maliga P, Dooner HK (1989) Visual detection of transposition of the maize element Activator (Ac) in tobacco seedlings. Science 244:204–207
Jones JDG, Carland F, Lim E, Ralston E, Donner HK (1990) Preferential transposition of the maize element Activator to linked chromosomal locations in tobacco. Plant Cell 2:701–707
Knapp S, Coupland G, Uhric H, Starlinger P, Salamini F (1988) Transposition of the maize transposable element Ac in Solanum tuberosum. Mol Gen Genet 213:285–290
Labarca C, Paigen K (1980) A simple, rapid, and sensitive DNA assay procedure. Anal Biochem 102:344–352
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Long D, Martin M, Swinburne J, Puangsomlee P, Coupland G (1993) The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: identification of an albino mutation induced by Ds insertion. Proc Natl Acad Sci USA 90:10370–10374
Mckenzie N, Wen LY, Dale PJ (2992) Tissue culture enhanced transposition of the maize transposable element Dissociation in Brassica oleracea var. italica. Theor Appl Genet 105:23–33 (2002)
Moloney MM, Walker JM, Sharma KK (1989) High-efficiency transformation of Brassica napus using Agrobacterium vectors. Plant Cell Rep 8:238–242
O’Neill C, Bancroft I (2000) Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J 23:233–243
Osborne BI, Corr CA, Prince JP, Hehel R, Tanksley SO, et al (1991) Ac transpositions from a T-DNA can generate linked and unlinked clusters of insertions in the tomato genome. Genetics 129:833–844
Parkin IAP, Sharpe AG, Keith DJ, Lydiate DJ (1995) Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38:1122–1131
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratroy Press, Cold Spring Harbor, N.Y.
Scofield SR, Harrison K, Nurrish SJ, Jones JDG (1992) Promoter fusions to the Activator transposase gene causes distinct patterns of Dissociation excision in tobacco cotyledons. Plant Cell 4:573–582
Scofield SR, Jones DA, Harrison K, Jones JDG (1994) Chloroplast targeting of spectinomycin adenyl transferase provides a cell-autonomous marker for monitoring transposon excision in tomato and tobacco. Mol Gen Genet 244:189–196
Sharp PJ, Kreis M, Shewry PR, Gale MD (1988) Location of β-amylase sequences in wheat and its relatives. Theor Appl Genet 75:286–290
Sharpe AG, Parkin IAP, Keith DJ, LydiateDJ (1995) Frequent nonreciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome 38:1112–1121
Springer PS, Mccombie WR, Sundaresan VA, Martienssen RA (1995) Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science 268:877–880
Swinburne J, Bacells L, Scofield SR, Jones JDG, Coupland G (1992) Elevated levels of Ac transposase mRNA are associated with high frequencies of Ds excision in Arabidopsis. Plant Cell 4:583–595
Szewc-McFadden AK, Kresovich S, Bliek SM, Mitchell SE, McFerson JR (1996) Identification of polymorphic, conserved simple sequence repeats (SSRs) in cultivated Brassica species. Theor Appl Genet 93:534–538
Thykjaer T, Stiller J, Handberg K, Jones J, Stougaard J (1995) The maize transposable element Ac is mobile in the legume Lotus japonicus. Plant Mol Biol 27:981–993
Van Sluys MA, Tempé J, Federoff N (1987) Transposition of the maize Activator element in Arabidopsis thaliana and Daucus carota. EMBO J 13:3881–3889
Vodkin LO (1989) Transposable element influence on plant gene expression and variation. Biochem Plants 15:83–133
Walbot V (1992) Strategies for mutagenesis and gene cloning using transposon tagging and T-DNA insertional mutagenesis. Annu Rev Plant Physiol Plant Mol Biol 43:49–82
Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C Baker B (1994) The product of the tobacco mosaic virus resistance gene N: Similarity to Toll and the Interleukin-1 receptor. Cell 78:1–20
Yoder JI, Pales J, Alpert K, Lassner M (1988) Ac transposition in transgenic tomato plants. Mol Gen Genet 213:219–296
Zhou JH, Atherly AG (1988) In situ detection of transposition of the maize controlling element Ac in transgenic soy bean tissues. Plant Cell Rep 8:542–545
Acknowledgements
The authors wish to thank Drs J. Jones, C. Dean and G. Coupland [John Innes Centre, Norwich, UK] for supplying the constructs, probe plasmids and also for providing details of the construction of plasmids. Special thanks to Penny Sparrow (JIC) for help with mapping data and analysis and also for maintaining plants used for construction of mapping lines. Thanks go also to Dr. Ian Bancroft, Dave Laurie and Eddie Arthur (JIC, Norwich) for critically reading the manuscript, as well as Martin Trick (JIC) for helpful advice throughout the project. This project was financially supported by the Biotechnology and Biological Sciences Research Council.
Author information
Authors and Affiliations
Additional information
Comunicated by C. Möllers
Rights and permissions
About this article
Cite this article
Mckenzie, N., Dale, P.J. Mapping of transposable element Dissociation inserts in Brassica oleracea following plant regeneration from streptomycin selection of callus. Theor Appl Genet 109, 333–341 (2004). https://doi.org/10.1007/s00122-004-1629-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1629-5