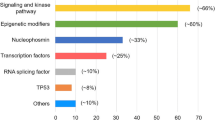
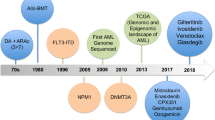
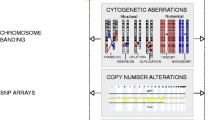
Abstract
Acute myeloid leukemia (AML) is an extremely heterogeneous disease defined by the clonal growth of myeloblasts/promyelocytes not only in the bone marrow but also in peripheral blood and/or tissues. Gene mutations and chromosomal abnormalities are usually associated with aberrant proliferation and/or block in the normal differentiation of hematopoietic cells. So far, the combination of cytogenetic profiling and molecular and gene mutation analyses remains an essential tool for the classification, diagnosis, prognosis, and treatment for AML. This review gives an overview on how the development of novel innovative technologies has allowed us not only to detect the genetic alterations as early as possible but also to understand the molecular pathogenesis of AML to develop novel targeted therapies. We also discuss the remarkable advances made during the last decade to understand the AML genome both at primary and relapse diseases and how genetic alterations might influence the distinct biological groups as well as the clonal evolution of disease during the diagnosis and relapse. Also, the review focuses on how the persistence of epigenetic gene mutations during morphological remission is associated with relapse. It is suggested that along with the prognostic and therapeutic mutations, the novel molecular targeted therapies either approved by FDA or those under clinical trials including CART-cell therapy would be of immense importance in the effective management of AML.



Similar content being viewed by others
Abbreviations
- AML:
-
acute myeloid leukemia
- APL:
-
acute promyelocytic leukemia
- ATRA:
-
all-trans-retinoic acid
- ATO:
-
arsenic trioxide
- ara-C:
-
cytosine arabinoside
- ASXL1/2:
-
additional sex Comb–like 1/2
- BCL-2:
-
B cell lymphoma 2 (BCL-2)
- BET:
-
bromodomain and extra-terminal
- BRD4:
-
bromodomain-containing protein 4
- CAR:
-
chimeric antigen receptor
- CBF:
-
core binding factor
- CEBPα:
-
CCAAT enhancer-binding protein alpha
- CRM1:
-
chromosomal maintenance 1
- CMP:
-
common myeloid progenitor
- CLP:
-
common lymphoid progenitor
- CMML:
-
chronic myelomonocytic leukemia
- CN-AML:
-
normal cytogenetic AML
- CR:
-
complete remission
- DNMT3A:
-
DNA methyltransferase 3 alpha
- ELN:
-
European Leukemia Net
- EZH2:
-
enhancer of zeste
- FAB:
-
French-American-British
- FDA:
-
Food and Drug Administration
- FLT3:
-
FMS-related tyrosine kinase 3
- 2-HG:
-
2-hydroxyglutarate
- HSC:
-
hematopoietic stem cell
- HSPC:
-
hematopoietic stem and progenitor cell
- Hh:
-
hedgehog
- IDH1/2:
-
isocitrate dehydrogenase 1/2
- ITD:
-
internal tandem duplication
- LSD1:
-
lysine-specific histone demethylase 1
- MRC:
-
Medical Research Council
- MDS:
-
myelodysplastic syndrome
- MLL-PTD:
-
mixed leukemia lineage-partial tandem duplication
- NPM1:
-
nucleophosmin
- NCCN:
-
National Comprehensive Cancer Network
- PLK1:
-
Polo-like kinase 1
- PDX:
-
patient-derived xenograft
- PML-RARA:
-
promyelocytic leukemia-retinoic acid receptor alpha
- RUNX1:
-
runt-related transcription factor 1
- SMC1:
-
structural maintenance of chromosomes 3
- STAG2:
-
stromal antigen 2
- TET2:
-
ten-eleven translocation 2
- TKD:
-
tyrosine kinase domain
- TP53:
-
tumor protein 53
- U2AF1:
-
U2 small nuclear RNA auxiliary factor
- VAF:
-
variant allele frequency
- WT1:
-
Wilms tumor 1
- WHO:
-
World Health Organization
- XPO1:
-
exportin-1
References
Estey E, Dohner H (2006) Acute myeloid leukaemia. Lancet 368:1894–1907
Lowenberg B, Downing JR, Burnett A (1999) Acute myeloid leukemia. N Engl J Med 341:1051–1062
Dohner H, Weisdorf DJ, Bloomfield CD (2015) Acute myeloid leukemia. N Engl J Med 373:1136–1152
Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ Jr, Laird PW et al (2013) Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368:2059–2074
Byrd JC, Mrozek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS et al (2002) Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood 100:4325–4336
Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA et al (2010) Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 115:453–474
Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S, Zang ZJ, Okuno Y, Shiraishi Y, Chiba K et al (2015) Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood 126:2491–2501
Othus M, Appelbaum FR, Petersdorf SH, Kopecky KJ, Slovak M, Nevill T, Brandwein J, Larson RA, Stiff PJ, Walter RB et al (2015) Fate of patients with newly diagnosed acute myeloid leukemia who fail primary induction therapy. Biol Blood Marrow Transplant 21:559–564
Schlenk RF (2014) Post-remission therapy for acute myeloid leukemia. Haematologica 99:1663–1670
Takami A (2018) Hematopoietic stem cell transplantation for acute myeloid leukemia. Int J Hematol 107:513–518
Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, Dombret H, Ebert BL, Fenaux P, Larson RA et al (2017) Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129:424–447
Mrozek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, Metzeler KH, Schwind S, Wu YZ, Kohlschmidt J et al (2012) Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 30:4515–4523
Gaidzik V, Dohner K (2008) Prognostic implications of gene mutations in acute myeloid leukemia with normal cytogenetics. Semin Oncol 35:346–355
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK, National Cancer Research Institute Adult Leukaemia Working G (2010) Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 116:354–365
Meyer SC, Levine RL (2014) Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol 15:e382–e394
Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C (1976) Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol 33:451–458
Vardiman JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellstrom-Lindberg E, Tefferi A et al (2009) The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 114:937–951
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW (2016) The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127:2391–2405
Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A et al (1998) The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood 92:2322–2333
Paschka P, Schlenk RF, Gaidzik VI, Herzig JK, Aulitzky T, Bullinger L, Spath D, Teleanu V, Kundgen A, Kohne CH et al (2015) ASXL1 mutations in younger adult patients with acute myeloid leukemia: a study by the German-Austrian Acute Myeloid Leukemia Study Group. Haematologica 100:324–330
Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M et al (2008) DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 456:66–72
Madan V, Shyamsunder P, Han L, Mayakonda A, Nagata Y, Sundaresan J, Kanojia D, Yoshida K, Ganesan S, Hattori N et al (2016) Comprehensive mutational analysis of primary and relapse acute promyelocytic leukemia. Leukemia 30:2430
Madan V, Shyamsunder P, Han L, Mayakonda A, Nagata Y, Sundaresan J, Kanojia D, Yoshida K, Ganesan S, Hattori N et al (2016) Comprehensive mutational analysis of primary and relapse acute promyelocytic leukemia. Leukemia 30:1672–1681
Chien W, Sun QY, Ding LW, Mayakonda A, Takao S, Liu L, Lim SL, Tan KT, Garg M, De Sousa Maria Varela A et al (2017) Diagnosis and relapse: cytogenetically normal acute myelogenous leukemia without FLT3-ITD or MLL-PTD. Leukemia 31:762–766
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N et al (2016) Genomic Classification and prognosis in acute myeloid leukemia. N Engl J Med 374:2209–2221
Bullinger L, Dohner K, Dohner H (2017) Genomics of acute myeloid leukemia diagnosis and pathways. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 35:934–946
Kadia TM, Ravandi F, Cortes J, Kantarjian H (2015) Toward individualized therapy in acute myeloid leukemia: A Contemporary Review. JAMA Oncol 1:820–828
Sun QY, Ding LW, Tan KT, Chien W, Mayakonda A, Lin DC, Loh XY, Xiao JF, Meggendorfer M, Alpermann T et al (2016) Ordering of mutations in acute myeloid leukemia with partial tandem duplication of MLL (MLL-PTD). Leukemia. https://doi.org/10.1038/leu.2016.160
Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M et al (2011) Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478:64–69
Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R et al (2013) Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 45:1232–1237
Cildir G, Pant H, Lopez AF, Tergaonkar V (2017) The transcriptional program, functional heterogeneity, and clinical targeting of mast cells. J Exp Med 214:2491–2506
Khattar E, Kumar P, Liu CY, Akincilar SC, Raju A, Lakshmanan M, Maury JJ, Qiang Y, Li S, Tan EY et al (2016) Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J Clin Invest 126:4045–4060
Thiede C, Koch S, Creutzig E, Steudel C, Illmer T, Schaich M, Ehninger G (2006) Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood 107:4011–4020
Schnittger S, Schoch C, Kern W, Mecucci C, Tschulik C, Martelli MF, Haferlach T, Hiddemann W, Falini B (2005) Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood 106:3733–3739
Falini B, Nicoletti I, Martelli MF, Mecucci C (2007) Acute myeloid leukemia carrying cytoplasmic/mutated nucleophosmin (NPMc+ AML): biologic and clinical features. Blood 109:874–885
Falini B, Bolli N, Shan J, Martelli MP, Liso A, Pucciarini A, Bigerna B, Pasqualucci L, Mannucci R, Rosati R et al (2006) Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood 107:4514–4523
Dohner K, Schlenk RF, Habdank M, Scholl C, Rucker FG, Corbacioglu A, Bullinger L, Frohling S, Dohner H (2005) Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood 106:3740–3746
Colombo E, Martinelli P, Zamponi R, Shing DC, Bonetti P, Luzi L, Volorio S, Bernard L, Pruneri G, Alcalay M et al (2006) Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res 66:3044–3050
Bolli N, De Marco MF, Martelli MP, Bigerna B, Pucciarini A, Rossi R, Mannucci R, Manes N, Pettirossi V, Pileri SA et al (2009) A dose-dependent tug of war involving the NPM1 leukaemic mutant, nucleophosmin, and ARF. Leukemia 23:501–509
Gu X, Ebrahem Q, Mahfouz RZ, Hasipek M, Enane F, Radivoyevitch T, Rapin N, Przychodzen B, Hu Z, Balusu R et al (2018) Leukemogenic nucleophosmin mutation disrupts the transcription factor hub that regulates granulomonocytic fates. J Clin Invest 128:4260–4279
Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O et al (2012) Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366:1079–1089
Estey EH (2018) Acute myeloid leukemia: 2019 update on risk-stratification and management. Am J Hematol 93:1267–1291
Cocciardi S, Dolnik A, Kapp-Schwoerer S, Rucker FG, Lux S, Blatte TJ, Skambraks S, Kronke J, Heidel FH, Schnoder TM et al (2019) Clonal evolution patterns in acute myeloid leukemia with NPM1 mutation. Nat Commun 10:2031
Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, Spath D, Kayser S, Zucknick M, Gotze K et al (2010) IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 28:3636–3643
Balusu R, Fiskus W, Rao R, Chong DG, Nalluri S, Mudunuru U, Ma H, Chen L, Venkannagari S, Ha K et al (2011) Targeting levels or oligomerization of nucleophosmin 1 induces differentiation and loss of survival of human AML cells with mutant NPM1. Blood 118:3096–3106
Perera Y, Farina HG, Gil J, Rodriguez A, Benavent F, Castellanos L, Gomez RE, Acevedo BE, Alonso DF, Perea SE (2009) Anticancer peptide CIGB-300 binds to nucleophosmin/B23, impairs its CK2-mediated phosphorylation, and leads to apoptosis through its nucleolar disassembly activity. Mol Cancer Ther 8:1189–1196
Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J et al (2010) DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363:2424–2433
Marcucci G, Metzeler KH, Schwind S, Becker H, Maharry K, Mrozek K, Radmacher MD, Kohlschmidt J, Nicolet D, Whitman SP et al (2012) Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 30:742–750
Gaidzik VI, Schlenk RF, Paschka P, Stolzle A, Spath D, Kuendgen A, von Lilienfeld-Toal M, Brugger W, Derigs HG, Kremers S et al (2013) Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG). Blood 121:4769–4777
Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S et al (2010) IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 28:2348–2355
Metzeler KH, Maharry K, Radmacher MD, Mrozek K, Margeson D, Becker H, Curfman J, Holland KB, Schwind S, Whitman SP et al (2011) TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29:1373–1381
Chou WC, Chou SC, Liu CY, Chen CY, Hou HA, Kuo YY, Lee MC, Ko BS, Tang JL, Yao M et al (2011) TET2 mutation is an unfavorable prognostic factor in acute myeloid leukemia patients with intermediate-risk cytogenetics. Blood 118:3803–3810
Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Gorlich D, Schneider S, Konstandin NP, Dufour A, Braundl K et al (2016) Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood 128:686–698
Shih AH, Abdel-Wahab O, Patel JP, Levine RL (2012) The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 12:599–612
Krivtsov AV, Armstrong SA (2007) MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer 7:823–833
Ernst P, Wang J, Korsmeyer SJ (2002) The role of MLL in hematopoiesis and leukemia. Curr Opin Hematol 9:282–287
Huret JL, Dessen P, Bernheim A (2001) An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia 15:987–989
Meyer C, Kowarz E, Hofmann J, Renneville A, Zuna J, Trka J, Ben Abdelali R, Macintyre E, De Braekeleer E, De Braekeleer M et al (2009) New insights to the MLL recombinome of acute leukemias. Leukemia 23:1490–1499
Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A et al (2010) Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42:722–726
Chase A, Cross NC (2011) Aberrations of EZH2 in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 17:2613–2618
Herviou L, Cavalli G, Cartron G, Klein B, Moreaux J (2016) EZH2 in normal hematopoiesis and hematological malignancies. Oncotarget 7:2284–2296
Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, Habdank M, Spath D, Morgan M, Benner A et al (2008) Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 358:1909–1918
Yu J, Li Y, Zhang D, Wan D, Jiang Z (2020) Clinical implications of recurrent gene mutations in acute myeloid leukemia. Exp Hematol Oncol 9:4
Gilliland DG, Griffin JD (2002) The roles of FLT3 in hematopoiesis and leukemia. Blood 100:1532–1542
Larrosa-Garcia M, Baer MR (2017) FLT3 Inhibitors in acute myeloid leukemia: current status and future directions. Mol Cancer Ther 16:991–1001
Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, Perl AE, Travers KJ, Wang S, Hunt JP et al (2012) Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 485:260–263
Pratz KW, Sato T, Murphy KM, Stine A, Rajkhowa T, Levis M (2010) FLT3-mutant allelic burden and clinical status are predictive of response to FLT3 inhibitors in AML. Blood 115:1425–1432
Gale RE, Green C, Allen C, Mead AJ, Burnett AK, Hills RK, Linch DC, Medical Research Council Adult Leukaemia Working P (2008) The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood 111:2776–2784
Sudhindra A, Smith CC (2014) FLT3 inhibitors in AML: are we there yet? Curr Hematol Malig Rep 9:174–185
Li Y, Cheng HS, Chng WJ, Tergaonkar V (2016) Activation of mutant TERT promoter by RAS-ERK signaling is a key step in malignant progression of BRAF-mutant human melanomas. Proc Natl Acad Sci U S A 113:14402–14407
Janssen JW, Steenvoorden AC, Lyons J, Anger B, Bohlke JU, Bos JL, Seliger H, Bartram CR (1987) RAS gene mutations in acute and chronic myelocytic leukemias, chronic myeloproliferative disorders, and myelodysplastic syndromes. Proc Natl Acad Sci U S A 84:9228–9232
Mrozek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD (2007) Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood 109:431–448
Wang CQ, Krishnan V, Tay LS, Chin DW, Koh CP, Chooi JY, Nah GS, Du L, Jacob B, Yamashita N et al (2014) Disruption of Runx1 and Runx3 leads to bone marrow failure and leukemia predisposition due to transcriptional and DNA repair defects. Cell Rep 8:767–782
Chakraborty S, Njah K, Pobbati AV, Lim YB, Raju A, Lakshmanan M, Tergaonkar V, Lim CT, Hong W (2017) Agrin as a mechanotransduction signal regulating YAP through the hippo pathway. Cell Rep 18:2464–2479
Meyers S, Downing JR, Hiebert SW (1993) Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol 13:6336–6345
Tang JL, Hou HA, Chen CY, Liu CY, Chou WC, Tseng MH, Huang CF, Lee FY, Liu MC, Yao M et al (2009) AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood 114:5352–5361
Marcucci G, Haferlach T, Dohner H (2011) Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29:475–486
Mendler JH, Maharry K, Radmacher MD, Mrozek K, Becker H, Metzeler KH, Schwind S, Whitman SP, Khalife J, Kohlschmidt J et al (2012) RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 30:3109–3118
Marcucci G, Mrozek K, Ruppert AS, Maharry K, Kolitz JE, Moore JO, Mayer RJ, Pettenati MJ, Powell BL, Edwards CG et al (2005) Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 23:5705–5717
Paschka P, Marcucci G, Ruppert AS, Mrozek K, Chen H, Kittles RA, Vukosavljevic T, Perrotti D, Vardiman JW, Carroll AJ et al (2006) Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 24:3904–3911
Park SH, Chi HS, Min SK, Park BG, Jang S, Park CJ (2011) Prognostic impact of c-KIT mutations in core binding factor acute myeloid leukemia. Leuk Res 35:1376–1383
Paschka P, Du J, Schlenk RF, Gaidzik VI, Bullinger L, Corbacioglu A, Spath D, Kayser S, Schlegelberger B, Krauter J et al (2013) Secondary genetic lesions in acute myeloid leukemia with inv(16) or t(16;16): a study of the German-Austrian AML Study Group (AMLSG). Blood 121:170–177
Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T et al (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149:214–231
Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M et al (2011) RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478:524–528
Dawson MA, Gudgin EJ, Horton SJ, Giotopoulos G, Meduri E, Robson S, Cannizzaro E, Osaki H, Wiese M, Putwain S et al (2014) Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia. Leukemia 28:311–320
Renner AG, Dos Santos C, Recher C, Bailly C, Creancier L, Kruczynski A, Payrastre B, Manenti S (2009) Polo-like kinase 1 is overexpressed in acute myeloid leukemia and its inhibition preferentially targets the proliferation of leukemic cells. Blood 114:659–662
Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K et al (2012) Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med 18:605–611
Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM et al (2012) The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21:473–487
Magliulo D, Bernardi R, Messina S (2018) Lysine-specific demethylase 1A as a promising target in acute myeloid leukemia. Front Oncol 8:255
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941–953
Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O’Dwyer KM et al (2013) BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 12:329–341
Zhou JD, Zhang TJ, Xu ZJ, Gu Y, Ma JC, Li XX, Guo H, Wen XM, Zhang W, Yang L et al (2019) BCL2 overexpression: clinical implication and biological insights in acute myeloid leukemia. Diagn Pathol 14:68
McMillan R, Matsui W (2012) Molecular pathways: the hedgehog signaling pathway in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 18:4883–4888
Lim Y, Gondek L, Li L, Wang Q, Ma H, Chang E, Huso DL, Foerster S, Marchionni L, McGovern K et al (2015) Integration of hedgehog and mutant FLT3 signaling in myeloid leukemia. Sci Transl Med 7:291–296
Cortes JE, Heidel FH, Hellmann A, Fiedler W, Smith BD, Robak T, Montesinos P, Pollyea DA, DesJardins P, Ottmann O et al (2019) Randomized comparison of low dose cytarabine with or without glasdegib in patients with newly diagnosed acute myeloid leukemia or high-risk myelodysplastic syndrome. Leukemia 33:379–389
Kojima K, Kornblau SM, Ruvolo V, Dilip A, Duvvuri S, Davis RE, Zhang M, Wang Z, Coombes KR, Zhang N et al (2013) Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood 121:4166–4174
Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M, Walker A, Klisovic R, Blum W, Caligiuri M et al (2012) Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood 120:1765–1773
Li Z, Guo JR, Chen QQ, Wang CY, Zhang WJ, Yao MC, Zhang W (2017) Exploring the antitumor mechanism of high-dose cytarabine through the metabolic perturbations of ribonucleotide and deoxyribonucleotide in human promyelocytic leukemia HL-60 cells. Molecules 22. https://doi.org/10.3390/molecules22030499
de The H, Chomienne C, Lanotte M, Degos L, Dejean A (1990) The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature 347:558–561
Greif PA, Yaghmaie M, Konstandin NP, Ksienzyk B, Alimoghaddam K, Ghavamzadeh A, Hauser A, Graf A, Krebs S, Blum H et al (2011) Somatic mutations in acute promyelocytic leukemia (APL) identified by exome sequencing. Leukemia 25:1519–1522
Kenderian SS, Ruella M, Shestova O, Klichinsky M, Aikawa V, Morrissette JJ, Scholler J, Song D, Porter DL, Carroll M et al (2015) CD33-specific chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia 29:1637–1647
Mardiros A, Dos Santos C, McDonald T, Brown CE, Wang X, Budde LE, Hoffman L, Aguilar B, Chang WC, Bretzlaff W et al (2013) T cells expressing CD123-specific chimeric antigen receptors exhibit specific cytolytic effector functions and antitumor effects against human acute myeloid leukemia. Blood 122:3138–3148
Brunetti L, Gundry MC, Sorcini D, Guzman AG, Huang YH, Ramabadran R, Gionfriddo I, Mezzasoma F, Milano F, Nabet B et al (2018) Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell 34(499-512):e499. https://doi.org/10.1016/j.ccell.2018.08.005
Takei H, Kobayashi SS (2019) Targeting transcription factors in acute myeloid leukemia. Int J Hematol 109:28–34
Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R (2009) Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 113:3088–3091
Pulikkan JA, Tenen DG, Behre G (2017) C/EBPalpha deregulation as a paradigm for leukemogenesis. Leukemia 31:2279–2285
Green CL, Koo KK, Hills RK, Burnett AK, Linch DC, Gale RE (2010) Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 28:2739–2747
Konstandin NP, Pastore F, Herold T, Dufour A, Rothenberg-Thurley M, Hinrichsen T, Ksienzyk B, Tschuri S, Schneider S, Hoster E et al (2018) Genetic heterogeneity of cytogenetically normal AML with mutations of CEBPA. Blood advances 2:2724–2731
Buscarlet M, Provost S, Zada YF, Bourgoin V, Mollica L, Dube MP, Busque L (2018) Lineage restriction analyses in CHIP indicate myeloid bias for TET2 and multipotent stem cell origin for DNMT3A. Blood 132:277–280
Grebien F, Vedadi M, Getlik M, Giambruno R, Grover A, Avellino R, Skucha A, Vittori S, Kuznetsova E, Smil D et al (2015) Pharmacological targeting of the Wdr5-MLL interaction in C/EBPalpha N-terminal leukemia. Nat Chem Biol 11:571–578
Chew CL, Conos SA, Unal B, Tergaonkar V (2018) Noncoding RNAs: Master regulators of inflammatory signaling. Trends Mol Med 24:66–84
Radomska HS, Jernigan F, Nakayama S, Jorge SE, Sun L, Tenen DG, Kobayashi SS (2015) A cell-based high-throughput screening for inducers of myeloid differentiation. J Biomol Screen 20:1150–1159
Sridhar R, Takei H, Syed R, Kobayashi IS, Hui LB, Kamal A, Tenen DG, Kobayashi SS (2018) Styryl quinazolinones as potential inducers of myeloid differentiation via upregulation of C/EBPalpha. Molecules 23. https://doi.org/10.3390/molecules23081938
Gaidzik VI, Bullinger L, Schlenk RF, Zimmermann AS, Rock J, Paschka P, Corbacioglu A, Krauter J, Schlegelberger B, Ganser A et al (2011) RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML study group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29:1364–1372
Haferlach T, Stengel A, Eckstein S, Perglerova K, Alpermann T, Kern W, Haferlach C, Meggendorfer M (2016) The new provisional WHO entity ‘RUNX1 mutated AML’ shows specific genetics but no prognostic influence of dysplasia. Leukemia 30:2109–2112
Khan M, Cortes J, Kadia T, Naqvi K, Brandt M, Pierce S, Patel KP, Borthakur G, Ravandi F, Konopleva M et al (2017) Clinical outcomes and co-occurring mutations in patients with RUNX1-mutated acute myeloid leukemia. International journal of molecular sciences 18. https://doi.org/10.3390/ijms18081618
Morita K, Suzuki K, Maeda S, Matsuo A, Mitsuda Y, Tokushige C, Kashiwazaki G, Taniguchi J, Maeda R, Noura M et al (2017) Genetic regulation of the RUNX transcription factor family has antitumor effects. J Clin Invest 127:2815–2828
Simon L, Lavallee VP, Bordeleau ME, Krosl J, Baccelli I, Boucher G, Lehnertz B, Chagraoui J, MacRae T, Ruel R et al (2017) Chemogenomic landscape of RUNX1-mutated AML reveals importance of RUNX1 allele dosage in genetics and glucocorticoid sensitivity. Clinical cancer research : an official journal of the American Association for Cancer Research 23:6969–6981
Egger G, Liang G, Aparicio A, Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429:457–463
Abdel-Wahab O, Levine RL (2013) Mutations in epigenetic modifiers in the pathogenesis and therapy of acute myeloid leukemia. Blood 121:3563–3572
Fong CY, Morison J, Dawson MA (2014) Epigenetics in the hematologic malignancies. Haematologica 99:1772–1783
Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW et al (2011) Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 43:309–315
Thol F, Damm F, Ludeking A, Winschel C, Wagner K, Morgan M, Yun H, Gohring G, Schlegelberger B, Hoelzer D et al (2011) Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29:2889–2896
Sehgal AR, Gimotty PA, Zhao J, Hsu JM, Daber R, Morrissette JD, Luger S, Loren AW, Carroll M (2015) DNMT3A Mutational status affects the results of dose-escalated induction therapy in acute myelogenous leukemia. Clinical cancer research : an official journal of the American Association for Cancer Research 21:1614–1620
Ball B, Zeidan A, Gore SD, Prebet T (2017) Hypomethylating agent combination strategies in myelodysplastic syndromes: hopes and shortcomings. Leuk Lymphoma 58:1022–1036
Metzeler KH, Walker A, Geyer S, Garzon R, Klisovic RB, Bloomfield CD, Blum W, Marcucci G (2012) DNMT3A mutations and response to the hypomethylating agent decitabine in acute myeloid leukemia. Leukemia 26:1106–1107
Cashen AF, Schiller GJ, O’Donnell MR, DiPersio JF (2010) Multicenter, phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 28:556–561
Issa JJ, Roboz G, Rizzieri D, Jabbour E, Stock W, O’Connell C, Yee K, Tibes R, Griffiths EA, Walsh K et al (2015) Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol 16:1099–1110
Kharfan-Dabaja MA (2015) Guadecitabine for AML and MDS: hype or hope? Lancet Oncol 16:1009–1011
Kantarjian H, Faderl S, Garcia-Manero G, Luger S, Venugopal P, Maness L, Wetzler M, Coutre S, Stock W, Claxton D et al (2012) Oral sapacitabine for the treatment of acute myeloid leukaemia in elderly patients: a randomised phase 2 study. Lancet Oncol 13:1096–1104
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A et al (2009) Mutation in TET2 in myeloid cancers. N Engl J Med 360:2289–2301
Weissmann S, Alpermann T, Grossmann V, Kowarsch A, Nadarajah N, Eder C, Dicker F, Fasan A, Haferlach C, Haferlach T et al (2012) Landscape of TET2 mutations in acute myeloid leukemia. Leukemia 26:934–942
Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R et al (2009) Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114:144–147
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF et al (2010) Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18:553–567
Gaidzik VI, Paschka P, Spath D, Habdank M, Kohne CH, Germing U, von Lilienfeld-Toal M, Held G, Horst HA, Haase D et al (2012) TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 30:1350–1357
Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG Jr (2013) (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339:1621–1625
Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE et al (2010) The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17:225–234
Yen K, Travins J, Wang F, David MD, Artin E, Straley K, Padyana A, Gross S, DeLaBarre B, Tobin E et al (2017) AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 MUTATIONS. Cancer Discov 7:478–493
Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, Stone RM, DeAngelo DJ, Levine RL, Flinn IW et al (2017) Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130:722–731
Pollyea DA, Tallman MS, de Botton S, Kantarjian HM, Collins R, Stein AS, Frattini MG, Xu Q, Tosolini A, See WL et al (2019) Enasidenib, an inhibitor of mutant IDH2 proteins, induces durable remissions in older patients with newly diagnosed acute myeloid leukemia. Leukemia. https://doi.org/10.1038/s41375-019-0472-2
Dohner K, Tobis K, Ulrich R, Frohling S, Benner A, Schlenk RF, Dohner H (2002) Prognostic significance of partial tandem duplications of the MLL gene in adult patients 16 to 60 years old with acute myeloid leukemia and normal cytogenetics: a study of the Acute Myeloid Leukemia Study Group Ulm. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 20:3254–3261
Caligiuri MA, Strout MP, Lawrence D, Arthur DC, Baer MR, Yu F, Knuutila S, Mrozek K, Oberkircher AR, Marcucci G et al (1998) Rearrangement of ALL1 (MLL) in acute myeloid leukemia with normal cytogenetics. Cancer Res 58:55–59
Kao HW, Liang DC, Kuo MC, Wu JH, Dunn P, Wang PN, Lin TL, Shih YS, Liang ST, Lin TH et al (2015) High frequency of additional gene mutations in acute myeloid leukemia with MLL partial tandem duplication: DNMT3A mutation is associated with poor prognosis. Oncotarget 6:33217–33225
Sun QY, Ding LW, Tan KT, Chien W, Mayakonda A, Lin DC, Loh XY, Xiao JF, Meggendorfer M, Alpermann T et al (2017) Ordering of mutations in acute myeloid leukemia with partial tandem duplication of MLL (MLL-PTD). Leukemia 31:1–10
Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, Allison DF, Cai L, Rockowitz S, Liu S et al (2015) Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood 125:346–357
Fujita S, Honma D, Adachi N, Araki K, Takamatsu E, Katsumoto T, Yamagata K, Akashi K, Aoyama K, Iwama A et al (2018) Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia 32:855–864
Basheer F, Giotopoulos G, Meduri E, Yun H, Mazan M, Sasca D, Gallipoli P, Marando L, Gozdecka M, Asby R et al (2019) Contrasting requirements during disease evolution identify EZH2 as a therapeutic target in AML. J Exp Med 216:966–981
Wingelhofer B, Somervaille TCP (2019) Emerging epigenetic therapeutic targets in acute myeloid leukemia. Front Oncol 9:850
Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, Kuscu C, Hricik T, Ndiaye-Lobry D, Lafave LM et al (2013) Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med 210:2641–2659
Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, Pandey S, Patel JP, Chung YR, Koche R et al (2012) ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 22:180–193
Metzeler KH, Becker H, Maharry K, Radmacher MD, Kohlschmidt J, Mrozek K, Nicolet D, Whitman SP, Wu YZ, Schwind S et al (2011) ASXL1 mutations identify a high-risk subgroup of older patients with primary cytogenetically normal AML within the ELN Favorable genetic category. Blood 118:6920–6929
Gelsi-Boyer V, Trouplin V, Adelaide J, Bonansea J, Cervera N, Carbuccia N, Lagarde A, Prebet T, Nezri M, Sainty D et al (2009) Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol 145:788–800
Huether R, Dong L, Chen X, Wu G, Parker M, Wei L, Ma J, Edmonson MN, Hedlund EK, Rusch MC et al (2014) The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun 5:3630
Yang H, Kurtenbach S, Guo Y, Lohse I, Durante MA, Li J, Li Z, Al-Ali H, Li L, Chen Z et al (2018) Gain of function of ASXL1 truncating protein in the pathogenesis of myeloid malignancies. Blood 131:328–341
Micol JB, Duployez N, Boissel N, Petit A, Geffroy S, Nibourel O, Lacombe C, Lapillonne H, Etancelin P, Figeac M et al (2014) Frequent ASXL2 mutations in acute myeloid leukemia patients with t(8;21)/RUNX1-RUNX1T1 chromosomal translocations. Blood 124:1445–1449
Carbuccia N, Trouplin V, Gelsi-Boyer V, Murati A, Rocquain J, Adelaide J, Olschwang S, Xerri L, Vey N, Chaffanet M et al (2010) Mutual exclusion of ASXL1 and NPM1 mutations in a series of acute myeloid leukemias. Leukemia 24:469–473
Liang DC, Liu HC, Yang CP, Jaing TH, Hung IJ, Yeh TC, Chen SH, Hou JY, Huang YJ, Shih YS et al (2013) Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood 121:2988–2995
Asada S, Goyama S, Inoue D, Shikata S, Takeda R, Fukushima T, Yonezawa T, Fujino T, Hayashi Y, Kawabata KC et al (2018) Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis. Nat Commun 9:2733
Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, Wang P, Yang H, Ma S, Lin H et al (2015) WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell 57:662–673
Paschka P, Marcucci G, Ruppert AS, Whitman SP, Mrozek K, Maharry K, Langer C, Baldus CD, Zhao W, Powell BL et al (2008) Wilms’ tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 26:4595–4602
Virappane P, Gale R, Hills R, Kakkas I, Summers K, Stevens J, Allen C, Green C, Quentmeier H, Drexler H et al (2008) Mutation of the Wilms’ tumor 1 gene is a poor prognostic factor associated with chemotherapy resistance in normal karyotype acute myeloid leukemia: the United Kingdom Medical Research Council Adult Leukaemia Working Party. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 26:5429–5435
Krauth MT, Alpermann T, Bacher U, Eder C, Dicker F, Ulke M, Kuznia S, Nadarajah N, Kern W, Haferlach C et al (2015) WT1 mutations are secondary events in AML, show varying frequencies and impact on prognosis between genetic subgroups. Leukemia 29:660–667
Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A et al (2014) DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep 9:1841–1855
Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T (2008) Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia 22:1539–1541
Prokocimer M, Molchadsky A, Rotter V (2017) Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: projections on diagnostic workup and therapy. Blood 130:699–712
Rucker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, Habdank M, Kugler CM, Holzmann K, Gaidzik VI et al (2012) TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood 119:2114–2121
Hou HA, Chou WC, Kuo YY, Liu CY, Lin LI, Tseng MH, Chiang YC, Liu MC, Liu CW, Tang JL et al (2015) TP53 mutations in de novo acute myeloid leukemia patients: longitudinal follow-ups show the mutation is stable during disease evolution. Blood Cancer J 5:e331. https://doi.org/10.1038/bcj.2015.59
Schlenk RF, Kayser S, Bullinger L, Kobbe G, Casper J, Ringhoffer M, Held G, Brossart P, Lubbert M, Salih HR et al (2014) Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood 124:3441–3449
Cortes JE, Kantarjian H, Foran JM, Ghirdaladze D, Zodelava M, Borthakur G, Gammon G, Trone D, Armstrong RC, James J et al (2013) Phase I study of quizartinib administered daily to patients with relapsed or refractory acute myeloid leukemia irrespective of FMS-like tyrosine kinase 3-internal tandem duplication status. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 31:3681–3687
Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini S, Montesinos P, Baer MR, Larson RA, Ustun C et al (2019) Gilteritinib or chemotherapy for relapsed or refractory FLT3-Mutated AML. N Engl J Med 381:1728–1740
Kadia TM, Kantarjian H, Kornblau S, Borthakur G, Faderl S, Freireich EJ, Luthra R, Garcia-Manero G, Pierce S, Cortes J et al (2012) Clinical and proteomic characterization of acute myeloid leukemia with mutated RAS. Cancer 118:5550–5559
Neubauer A, Maharry K, Mrozek K, Thiede C, Marcucci G, Paschka P, Mayer RJ, Larson RA, Liu ET, Bloomfield CD (2008) Patients with acute myeloid leukemia and RAS mutations benefit most from postremission high-dose cytarabine: a Cancer and Leukemia Group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 26:4603–4609
Maiti A, Naqvi K, Kadia TM, Borthakur G, Takahashi K, Bose P, Daver NG, Patel A, Alvarado Y, Ohanian M et al (2019) Phase II trial of MEK inhibitor binimetinib (MEK162) in RAS-mutant acute myeloid leukemia. Clin Lymphoma Myeloma Leuk 19:142-148 e141
Sattler M, Salgia R (2004) Targeting c-Kit mutations: basic science to novel therapies. Leuk Res 28(Suppl 1):S11–S20
Jung CL, Kim HJ, Kim DH, Huh H, Song MJ, Kim SH (2011) CKIT mutation in therapy-related acute myeloid leukemia with MLLT3/MLL chimeric transcript from t(9;11)(p22;q23). Ann Clin Lab Sci 41:193–196
Allen C, Hills RK, Lamb K, Evans C, Tinsley S, Sellar R, O’Brien M, Yin JL, Burnett AK, Linch DC et al (2013) The importance of relative mutant level for evaluating impact on outcome of KIT, FLT3 and CBL mutations in core-binding factor acute myeloid leukemia. Leukemia 27:1891–1901
Kanojia D, Garg M, Martinez J, M TA, Luty SB, Doan NB, Said JW, Forscher C, Tyner JW, Koeffler HP (2017) Kinase profiling of liposarcomas using RNAi and drug screening assays identified druggable targets. J Hematol Oncol 10:173
Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H et al (2007) Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3:576–583
Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y (2007) Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3:570–575
Seiler M, Yoshimi A, Darman R, Chan B, Keaney G, Thomas M, Agrawal AA, Caleb B, Csibi A, Sean E et al (2018) H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat Med 24:497–504
Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J et al (2012) The origin and evolution of mutations in acute myeloid leukemia. Cell 150:264–278
Wartman LD, Larson DE, Xiang Z, Ding L, Chen K, Lin L, Cahan P, Klco JM, Welch JS, Li C et al (2011) Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest 121:1445–1455
Ibanez M, Carbonell-Caballero J, Garcia-Alonso L, Such E, Jimenez-Almazan J, Vidal E, Barragan E, Lopez-Pavia M, M LL, Martin I et al (2016) The mutational landscape of acute promyelocytic leukemia reveals an interacting network of co-occurrences and recurrent mutations. PLoS One 11:e0148346. https://doi.org/10.1371/journal.pone.0148346
Swaminathan S, Garg S, Madkaikar M, Gupta M, Jijina F, Ghosh K (2014) FLT3 and NPM-1 mutations in a cohort of acute promyelocytic leukemia patients from India. Indian J Hum Genet 20:160–165
Wang ZY, Chen Z (2008) Acute promyelocytic leukemia: from highly fatal to highly curable. Blood 111:2505–2515
Shen ZX, Shi ZZ, Fang J, Gu BW, Li JM, Zhu YM, Shi JY, Zheng PZ, Yan H, Liu YF et al (2004) All-trans retinoic acid/As2O3 combination yields a high quality remission and survival in newly diagnosed acute promyelocytic leukemia. Proc Natl Acad Sci U S A 101:5328–5335
Ravandi F, Estey E, Jones D, Faderl S, O’Brien S, Fiorentino J, Pierce S, Blamble D, Estrov Z, Wierda W et al (2009) Effective treatment of acute promyelocytic leukemia with all-trans-retinoic acid, arsenic trioxide, and gemtuzumab ozogamicin. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 27:504–510
Passegue E, Jamieson CH, Ailles LE, Weissman IL (2003) Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci U S A 100(Suppl 1):11842–11849
Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R (2014) Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A 111:2548–2553
Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD et al (2012) Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481:506–510
Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, Majeti R (2012) Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med 4:149ra118
Ploen GG, Nederby L, Guldberg P, Hansen M, Ebbesen LH, Jensen UB, Hokland P, Aggerholm A (2014) Persistence of DNMT3A mutations at long-term remission in adult patients with AML. Br J Haematol 167:478–486
Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S, Wartman LD, Christopher M, Lamprecht TL, Helton NM et al (2015) Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA 314:811–822
Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW et al (2014) Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506:328–333
Jan M, Ebert BL, Jaiswal S (2017) Clonal hematopoiesis. Semin Hematol 54:43–50
Koeffler HP, Leong G (2017) Preleukemia: one name, many meanings. Leukemia 31:534–542
Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A et al (2014) Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371:2488–2498
Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA et al (2014) Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20:1472–1478
Lancet JE, Cortes JE, Hogge DE, Tallman MS, Kovacsovics TJ, Damon LE, Komrokji R, Solomon SR, Kolitz JE, Cooper M et al (2014) Phase 2 trial of CPX-351, a fixed 5:1 molar ratio of cytarabine/daunorubicin, vs cytarabine/daunorubicin in older adults with untreated AML. Blood 123:3239–3246
Lancet JE, Uy GL, Cortes JE, Newell LF, Lin TL, Ritchie EK, Stuart RK, Strickland SA, Hogge D, Solomon SR et al (2018) CPX-351 (cytarabine and daunorubicin) liposome for injection versus conventional cytarabine plus daunorubicin in older patients with newly diagnosed secondary acute myeloid leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 36:2684–2692
Rudolph D, Impagnatiello MA, Blaukopf C, Sommer C, Gerlich DW, Roth M, Tontsch-Grunt U, Wernitznig A, Savarese F, Hofmann MH et al (2015) Efficacy and mechanism of action of volasertib, a potent and selective inhibitor of Polo-like kinases, in preclinical models of acute myeloid leukemia. J Pharmacol Exp Ther 352:579–589
Gutteridge RE, Ndiaye MA, Liu X, Ahmad N (2016) Plk1 inhibitors in cancer therapy: from laboratory to clinics. Mol Cancer Ther 15:1427–1435
Lee KS, Burke TR Jr, Park JE, Bang JK, Lee E (2015) Recent advances and new strategies in targeting Plk1 for anticancer therapy. Trends Pharmacol Sci 36:858–877
Janning M, Fiedler W (2014) Volasertib for the treatment of acute myeloid leukemia: a review of preclinical and clinical development. Future Oncol 10:1157–1165
Moison C, Lavallee VP, Thiollier C, Lehnertz B, Boivin I, Mayotte N, Gareau Y, Frechette M, Blouin-Chagnon V, Corneau S et al (2019) Complex karyotype AML displays G2/M signature and hypersensitivity to PLK1 inhibition. Blood advances 3:552–563
Sprussel A, Schulte JH, Weber S, Necke M, Handschke K, Thor T, Pajtler KW, Schramm A, Konig K, Diehl L et al (2012) Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia 26:2039–2051
Smitheman KN, Severson TM, Rajapurkar SR, McCabe MT, Karpinich N, Foley J, Pappalardi MB, Hughes A, Halsey W, Thomas E et al (2019) Lysine specific demethylase 1 inactivation enhances differentiation and promotes cytotoxic response when combined with all-trans retinoic acid in acute myeloid leukemia across subtypes. Haematologica 104:1156–1167
Abdel-Aziz AK, Pallavicini I, Ceccacci E, Meroni G, Saadeldin MK, Varasi M, Minucci S (2019) Tuning mTORC1 activity dictates the response of acute myeloid leukemia to LSD1 inhibition. Haematologica. https://doi.org/10.3324/haematol.2019.224501
DiNardo CD, Pratz K, Pullarkat V, Jonas BA, Arellano M, Becker PS, Frankfurt O, Konopleva M, Wei AH, Kantarjian HM et al (2019) Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood 133:7–17
Wei AH, Strickland SA Jr, Hou JZ, Fiedler W, Lin TL, Walter RB, Enjeti A, Tiong IS, Savona M, Lee S et al (2019) Venetoclax combined with low-dose cytarabine for previously untreated patients with acute myeloid leukemia: results from a phase Ib/II study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 37:1277–1284
Garg M, Kanojia D, Mayakonda A, Ganesan TS, Sadhanandhan B, Suresh S, S S, Nagare RP, Said JW, Doan NB et al (2017) Selinexor (KPT-330) has antitumor activity against anaplastic thyroid carcinoma in vitro and in vivo and enhances sensitivity to doxorubicin. Sci Rep 7:9749
Garg M, Kanojia D, Mayakonda A, Said JW, Doan NB, Chien W, Ganesan TS, Chuang LS, Venkatachalam N, Baloglu E et al (2017) Molecular mechanism and therapeutic implications of selinexor (KPT-330) in liposarcoma. Oncotarget 8:7521–7532
Talati C, Sweet KL (2018) Nuclear transport inhibition in acute myeloid leukemia: recent advances and future perspectives. Int J Hematol Oncol 7:IJH04
Etchin J, Berezovskaya A, Conway AS, Galinsky IA, Stone RM, Baloglu E, Senapedis W, Landesman Y, Kauffman M, Shacham S et al (2016) KPT-8602, a second-generation inhibitor of XPO1-mediated nuclear export, is well tolerated and highly active against AML blasts and leukemia-initiating cells. Leukemia. https://doi.org/10.1038/leu.2016.145
Etchin J, Berezovskaya A, Conway AS, Galinsky IA, Stone RM, Baloglu E, Senapedis W, Landesman Y, Kauffman M, Shacham S et al (2017) KPT-8602, a second-generation inhibitor of XPO1-mediated nuclear export, is well tolerated and highly active against AML blasts and leukemia-initiating cells. Leukemia 31:143–150
Jackson HJ, Rafiq S, Brentjens RJ (2016) Driving CAR T-cells forward. Nat Rev Clin Oncol 13:370–383
Warrier VU, Makandar AI, Garg M, Sethi G, Kant R, Pal JK, Yuba E, Gupta RK (2019) Engineering anti-cancer nanovaccine based on antigen cross-presentation. Biosci Rep:39. https://doi.org/10.1042/BSR20193220
Brentjens RJ, Riviere I, Park JH, Davila ML, Wang X, Stefanski J, Taylor C, Yeh R, Bartido S, Borquez-Ojeda O et al (2011) Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood 118:4817–4828
Kochenderfer JN, Rosenberg SA (2013) Treating B-cell cancer with T cells expressing anti-CD19 chimeric antigen receptors. Nat Rev Clin Oncol 10:267–276
Ross JF, Wang H, Behm FG, Mathew P, Wu M, Booth R, Ratnam M (1999) Folate receptor type beta is a neutrophilic lineage marker and is differentially expressed in myeloid leukemia. Cancer 85:348–357
Lynn RC, Poussin M, Kalota A, Feng Y, Low PS, Dimitrov DS, Powell DJ Jr (2015) Targeting of folate receptor beta on acute myeloid leukemia blasts with chimeric antigen receptor-expressing T cells. Blood 125:3466–3476
Casucci M, Nicolis di Robilant B, Falcone L, Camisa B, Norelli M, Genovese P, Gentner B, Gullotta F, Ponzoni M, Bernardi M et al (2013) CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood 122:3461–3472
Funding
This work was supported and funded by the Department of Biotechnology (DBT), Government of India under its Ramalingaswami Fellowship (no. BT/RLF/Re-entry/24/2014) award to Dr. Manoj Garg. Science & Engineering Research Board (SERB; ECR/2016/001519), DST, Government of India. The figures were drawn by using BioRender online software (https://biorender.com).
Author information
Authors and Affiliations
Contributions
MG conceived the idea and designed the format of the manuscript. MG, AK, GP, AKP, GS, and BCD have organized and presented the concepts in the manuscript. MG, AK, and GP created the figures and tables. MG and BCD revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kirtonia, A., Pandya, G., Sethi, G. et al. A comprehensive review of genetic alterations and molecular targeted therapies for the implementation of personalized medicine in acute myeloid leukemia. J Mol Med 98, 1069–1091 (2020). https://doi.org/10.1007/s00109-020-01944-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-020-01944-5