Abstract
KIBRA has been described as a key regulator of the Hippo signaling pathway, regulating organ size control, cell contact inhibition, cell growth, as well as tumorigenesis and cystogenesis. Since there is scarce information on KIBRA gene expression regulation, we analyzed the molecular basis of tissue-specific KIBRA expression in human kidney epithelial (IHKE, HPCT) and neuroblastoma (SH-SY5Y, SK-SN-SH) cells. We detected four novel and differentially used transcription start sites, two of which positioned in the first intron, generating two novel alternative exons. We identified one constitutively active core promoter (P1a) and three alternative promoters (P1b, P2, and P3), which were exclusively active in kidney cells. Transcription factor 7-like 2 (TCF7L2) selectively activated KIBRA at P1a, P2, and P3 in kidney cells. The two genetic variants −580C>T (p < 0.05) and −1691C>T (p < 0.01) significantly affected the transcriptional activity of the KIBRA core promoter. We propose a novel functional structure of the KIBRA gene and provide detailed insight into molecular cell type-specific KIBRA transcriptional regulation by TCF7L2, the Yes-associated protein 1 and TEA domain family member. Our findings provide a potential basis for future studies on malfunctioning KIBRA regulation in pathophysiological conditions such as cancer development.
Key message
-
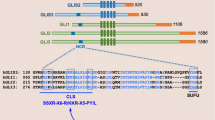
KIBRA expression is regulated by three independent, cell type-specific promoters
-
Two novel TSS were located within intron one resulting in two alternative exons
-
TSS utilization is cell type-specific
-
TCF7L2, YAP1, and TEAD are involved in the differential KIBRA expression regulation







Similar content being viewed by others

References
Kremerskothen J, Plaas C, Büther K, Finger I, Veltel S, Matanis T, Liedtke T, Barnekow A (2003) Characterization of KIBRA, a novel WW domain-containing protein. Biochem Biophys Res Commun 300:862–867
Almeida OP, Schwab SG, Lautenschlager NT, Morar B, Greenop KR, Flicker L, Wildenauer D (2008) KIBRA genetic polymorphism influences episodic memory in later life, but does not increase the risk of mild cognitive impairment. J Cell Mol Med 12:1672–1676
Bates TC, Price JF, Harris SE, Marioni RE, Fowkes FG, Stewart MC, Murray GD, Whalley LJ, Starr JM, Deary IJ (2009) Association of KIBRA and memory. Neurosci Lett 458:140–143
Nacmias B, Bessi V, Bagnoli S, Tedde A, Cellini E, Piccini C, Sorbi S, Bracco L (2008) KIBRA gene variants are associated with episodic memory performance in subjective memory complaints. Neurosci Lett 436:145–147
Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D et al (2006) Common Kibra alleles are associated with human memory performance. Science 314:475–478
Schaper K, Kolsch H, Popp J, Wagner M, Jessen F (2008) KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging 29:1123–1125
Schneider A, Huentelman MJ, Kremerskothen J, Duning K, Spoelgen R, Nikolich K (2010) KIBRA: a new gateway to learning and memory? Front Aging Neurosci 2:4
Wersching H, Guske K, Hasenkamp S, Hagedorn C, Schiwek S, Jansen S, Witte V, Wellmann J, Lohmann H, Duning K et al (2011) Impact of common KIBRA allele on human cognitive functions. Neuropsychopharmacology 36:1296–1304
Burgess JD, Pedraza O, Graff-Radford NR, Hirpa M, Zou F, Miles R, Nguyen T, Li M, Lucas JA, Ivnik RJ et al (2011) Association of common KIBRA variants with episodic memory and AD risk. Neurobiol Aging 32:557.e1–557.e9
Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Craig DW et al (2010) Evidence for an association between KIBRA and late-onset Alzheimer's disease. Neurobiol Aging 31:901–909
Rodríguez-Rodríguez E, Infante J, Llorca J, Mateo I, Sánchez-Quintana C, García-Gorostiaga I, Sánchez-Juan P, Berciano J, Combarros O (2009) Age-dependent association of KIBRA genetic variation and Alzheimer's disease risk. Neurobiol Aging 30:322–324
Büther K, Plaas C, Barnekow A, Kremerskothen J (2004) KIBRA is a novel substrate for protein kinase Czeta. Biochem Biophys Res Commun 317:703–707
Duning K, Schurek EM, Schlüter M, Bayer M, Reinhardt HC, Schwab A, Schaefer L, Benzing T, Schermer B, Saleem MA et al (2008) KIBRA modulates directional migration of podocytes. J Am Soc Nephrol 19:1891–1903
Kremerskothen J, Kindler S, Finger I, Veltel S, Barnekow A (2006) Postsynaptic recruitment of Dendrin depends on both dendritic mRNA transport and synaptic anchoring. J Neurochem 96:1659–1666
Mundel P, Heid HW, Mundel TM, Krüger M, Reiser J, Kriz W (1997) Synaptopodin: an actin-associated protein in telencephalic dendrites and renal podocytes. J Cell Biol 139:193–204
Rosse C, Formstecher E, Boeckeler K, Zhao Y, Kremerskothen J, White MD, Camonis JH, Parker PJ (2009) An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLoS Biol 7:e1000235
Yoshihama Y, Sasaki K, Horikoshi Y, Suzuki A, Ohtsuka T, Hakuno F, Takahashi S, Ohno S, Chida K (2011) KIBRA suppresses apical exocytosis through inhibition of aPKC kinase activity in epithelial cells. Curr Biol 21:705–711
Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H (2010) The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell 18:309–316
Genevet A, Tapon N (2011) The Hippo pathway and apico-basal cell polarity. Biochem J 436:213–224
Xiao L, Chen Y, Ji M, Dong J (2011) KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J Biol Chem 286:7788–7796
Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D (2010) Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 18:288–299
Boggiano JC, Fehon RG (2012) Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev Cell 22:695–702
Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G, Pandolfi PP, Givol D, Strano S, Lu X et al (2008) PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol Cell 32:803–814
Liu H, Jiang D, Chi F, Zhao B (2012) The Hippo pathway regulates stem cell proliferation, self-renewal, and differentiation. Protein Cell 3:291–304
Moleirinho S, Chang N, Sims AH, Tilston-Lünel AM, Angus L, Steele A, Boswell V, Barnett SC, Ormandy C, Faratian D et al (2012) KIBRA exhibits MST-independent functional regulation of the Hippo signaling pathway in mammals. Oncogene. http://www.nature.com/onc/journal/vaop/ncurrent/full/onc2012196a.html
Happé H, de Heer H, Peters DJ (2011) Polycystic kidney disease: the complexity of planar cell polarity and signaling during tissue regeneration and cyst formation. Biochim Biophys Acta 1812:1249–1255
Hohage H, Stachon A, Feidt C, Hirsch JR, Schlatter E (1998) Regulation of organic cation transport in IHKE-1 and LLC-PK1 cells. Fluorometric studies with 4-(4-dimethylaminostyryl)-N-methylpyridinium. J Pharmacol Exp Ther 286:305–310
Jessen H, Røigaard H, Riahi-Esfahani S, Jacobsen C (1994) A comparative study on the uptake of alpha-aminoisobutyric acid by normal and immortalized human embryonic kidney cells from proximal tubule. Biochim Biophys Acta 1190:279–288
Tveito G, Hansteen IL, Dalen H, Haugen A (1989) Immortalization of normal human kidney epithelial cells by nickel (II). Cancer Res 49:1829–1835
Orosz DE, Woost PG, Kolb RJ, Finesilver MB, Jin W, Frisa PS, Choo CK, Yau CF, Chan KW, Resnick MI et al (2004) Growth, immortalization, and differentiation potential of normal adult human proximal tubule cells. In Vitro Cell Dev Biol Anim 40:22–34
Liao Y, Lönnerdal B (2010) beta-Catenin/TCF4 transactivates miR-30e during intestinal cell differentiation. Cell Mol Life Sci 67:2969–2978
Schaefer BC (1995) Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem 227:255–273
Dördelmann C, Telgmann R, Brand E, Hagedorn C, Schröer B, Hasenkamp S, Baumgart P, Kleine-Katthöfer P, Paul M, Brand-Herrmann SM (2008) Functional and structural profiling of the human thrombopoietin gene promoter. J Biol Chem 283:24382–24391
Grabe N (2002) AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol 2:S1–S15
Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18:333–334
Schmitz B, Salomon A, Rötrige A, Ritter M, Ringelstein EB, Fischer JW, Paul M, Brand E, Brand SM (2013) Interindividual transcriptional regulation of the human biglycan gene involves three common molecular haplotypes. Arterioscler Thromb Vasc Biol 33:871–880
Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC et al (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38:626–635
Hochheimer A, Tjian R (2003) Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev 17:1309–1320
Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B (2005) A high-resolution map of active promoters in the human genome. Nature 436:876–880
Lee MP, Howcroft K, Kotekar A, Yang HH, Buetow KH, Singer DS (2005) ATG deserts define a novel core promoter subclass. Genome Res 15:1189–1197
Tsuchihara K, Suzuki Y, Wakaguri H, Irie T, Tanimoto K, Hahimoto S, Matsushima K, Mizushima-Sugano J, Yamashita R, Nakai K et al (2009) Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res 37:2249–2263
Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ et al (2003) The UCSC genome browser database. Nucleic Acids Res 31:51–54
Thomas DJ, Rosenbloom KR, Clawson H, Hinrichs AS, Trumbower H, Raney BJ, Karolchik D, Barber GP, Harte RA, Hillman-Jackson J et al (2007) The ENCODE Project at UC Santa Cruz. Nucleic Acids Res 35:D663–D667
Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845–1848
Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J et al (2007) Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res 17:746–759
Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA (2008) Divergent transcription from active promoters. Science 322:1849–1851
Kruglyak L, Nickerson DA (2001) Variation is the spice of life. Nat Genet 27:234–236
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Zhao J, Schug J, Li M, Kaestner KH, Grant SF (2010) Disease-associated loci are significantly over-represented among genes bound by transcription factor 7like 2 (TCF7L2) in vivo. Diabetologia 53:2340–2346
Brabletz T, Jung A, Dag S, Hlubek F, Kirchner T (1999) beta-Catenin regulates the expression of the matrix metalloproteinase-7 in human colorectal cancer. Am J Pathol 155:1033–1038
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW (1998) Identification of c-MYC as a target of the APC pathway. Science 281:1509–1512
Tetsu O, McCormick F (1999) beta-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422–426
Smale ST, Kadonaga JT (2003) The RNA polymerase II core promoter. Annu Rev Biochem 72:449–479
Kozak M (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44:283–292
Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H (1998) Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol Cell Biol 18:1248–1256
Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H (1998) Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 19:379–383
Li J, Sutter C, Parker DS, Blauwkamp T, Fang M, Cadigan KM (2007) CBP/p300 are bimodal regulators of Wnt signaling. EMBO J 26:2284–2294
Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H et al (2009) HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci 12:829–838
Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF (2011) Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332:458–461
Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N (2010) Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell 18:300–308
Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM et al (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22:1962–1971
Roose J, Clevers H (1999) TCF transcription factors: molecular switches in carcinogenesis. Biochim Biophys Acta 1424:M23–M37
Acknowledgments
We are grateful to Peter Kleine-Katthöfer (St. Franziskus-Hospital Münster) and Peter Baumgart (Clemenshospital GmbH Münster), supplying genomic DNA for the MolProMD Study. IHKE and HPCT cells were a kind gift of Eberhardt Schlatter, Department of Experimental Nephrology, University Hospital Münster and Ulrich Hopfer, Department of Physiology and Biophysics, Case Western Reserve University School of Medicine. Human brain cortex tissue was provided by Tanja Kuhlmann, Institute of Neuropathology, University Hospital Münster. We thank Birgit Orlowski and Alois Rötrige for excellent technical assistance. Eva Brand is supported by a Heisenberg professorship from the Deutsche Forschungsgemeinschaft (Br1589/8-2). This study was also supported by a grant from the European Union-Project Network of Excellence, FP6-2005-LIFESCIHEALTH-6, Integrating Genomics, Clinical Research and Care in Hypertension, InGenious HyperCare (proposal no. 037093 to Eva Brand and Stefan-Martin Brand) and an ICT in the FP7-ICT-2007-2, project number 224635, VPH2-Virtual Pathological Heart of the Virtual Physiological Human (supported Boris Schmitz), to Stefan-Martin Brand, formerly Stefan-Martin BrandHerrmann/Stefan-Martin Herrmann.
Conflict of interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 511 kb)
Rights and permissions
About this article
Cite this article
Guske, K., Schmitz, B., Schelleckes, M. et al. Tissue-specific differences in the regulation of KIBRA gene expression involve transcription factor TCF7L2 and a complex alternative promoter system. J Mol Med 92, 185–196 (2014). https://doi.org/10.1007/s00109-013-1089-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-013-1089-y