Abstract
Estuaries, being transition zones between land and ocean, act as sink or source of nitrate and thus influence the conditions in adjacent coastal waters. Hence, nitrification, which is the process oxidizing ammonium via nitrite to nitrate and simultaneously consuming oxygen, is important in estuaries. The process has been studied in sediment and water column of many estuaries, but seldom in both estuarine compartments at the same time. In August 2014, we collected water and sediment samples during a sampling trip along the salinity gradient of the hyper-turbid Ems estuary, which ends up in the North Sea. We conducted nitrification incubations in microcosms to determine nitrification potentials and we measured a suite of abiotic factors like oxygen saturation, salinity, and dissolved inorganic nitrogen (DIN). Two approaches were used, one isotope dilution method for net (NNP) and gross (GNP) nitrification potentials and one method with substrate addition for substrate induced nitrification potentials (SNP). The long-term incubation set-ups of several days include inseparably nitrification-coupled processes like remineralization and nitrate consumption, as well as cell growth, and hence they do not represent in-situ rates of nitrification. DNA was also isolated and used for quantitative PCR of the archaeal and bacterial amoA genes, which encode for the ammonia-oxidizing enzyme ammonia monooxygenase (AMO). Nitrification varied over the salinity gradient of the estuary. GNP in water and sediments decreased with increasing salinity. No NNP could be measured in the sediments of the oligohaline part of the estuary, while SNP was four-fold higher than GNP in this part of the estuary. Generally, the gene abundance of the amoA gene was higher in the oligohaline/mesohaline area than in the polyhaline area, and archaea dominated the ammonia-oxidizing communities in all samples. The local similarity in partitioning of archaeal and bacterial amoA genes over the water column and sediment at each sampling station along the estuarine gradient implied a link between the archaeal and bacterial ammonia oxidizers in both compartments, which is likely due to resuspension of sediment particles in the water column of this hyper-turbid estuary.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Estuaries play a major role in transport of nutrients and organic matter from the land to the coastal zones. It is commonly assumed that they have a filter function due to high turnover capacities (Crossland et al. 2005). Depending on the conditions in the estuary like the amount of available oxygen, the load of dissolved inorganic nitrogen or the quantity of suspended matter in the water column, estuaries can act as a source or a sink of nitrate (Dähnke et al. 2008; Middelburg and Nieuwenhuize 2001). The load of nitrate transported to coastal zones is dependent on the net effect of the microbial processes of nitrification and denitrification converting reactive nitrogen pools from anthropogenic inputs (Galloway et al. 2003). This can result in the eutrophication of coastal waters (Howarth and Marino 2006) as has happened for example in the North Sea (Radach and Pätsch 2007). Due to better waste water management and control of fertilizers, the riverine load of reactive nitrogen into the North Sea has decreased but is still high (Pätsch et al. 2010).
Along the estuaries, gradients of salinity, nitrogen, pH and oxygen are commonly formed and some of these variables have been shown to be influenced by or impacting microbial communities (Bernhard et al. 2005a) including nitrifiers (Bernhard and Bollmann 2010) and specially the ammonia oxidizers (Bernhard et al. 2005b, 2007; Mosier and Francis 2008; Santoro et al. 2008). Despite the obvious ecological importance of nitrification, the role of environmental factors that drive the associated communities of ammonia oxidizers is poorly understood. Niche differentiation between the two major groups of ammonia oxidizers, i.e. the ammonia-oxidizing archaea and bacteria is still under debate. The functional amoA gene encodes the active site of the ammonia monooxygenase that is specific for ammonia-oxidizing microorganisms. Recently the debate expanded with the discovery of nitrite-oxidizing bacteria that are able to carry out the whole process of nitrification including ammonia oxidation (Daims et al. 2015; van Kessel et al. 2015). There are many studies on nitrification in estuaries around the world, but most of these studies have either focused on sediments (Damashek et al. 2015; Mosier and Francis 2008; Sahan and Muyzer 2008; Wankel et al. 2011) or on water column (Bollmann and Laanbroek 2002; Damashek et al. 2016; de Bie et al. 2001; Heiss and Fulweiler 2016; Zhang et al. 2015). Studies that deal with both compartments of estuaries are apparently lacking, while such studies seem to be essential in well-mixed, shallow river mouths. For the determination of nitrification a number of different approaches exist and have been used (Damashek et al. 2016 and references therein). The most common method in coastal and ocean waters uses 15N-labelled ammonium in short-term incubations for 6–48 h. This method presents most likely the in-situ activity without growth of nitrifiers and without much influence of other processes like remineralization. Due to apparently low in-situ nitrification rates, we have chosen different incubation set-ups with a longer incubation time. In one of the set-ups, we applied an isotope dilution approach by labeling the end-product of nitrification nitrate, in the other set-up we added ammonium as substrate. The nitrification potential obtained in long-term incubations include ammonification of organic matter and nitrate loss by e.g. denitrification, as well as changes in nitrifier community and growth. Consequently, the results are not direct comparable with other studies and the measured rates could be overestimations of in-situ rates. However, they should represent a range of nitrification potentials in the Ems estuary related to the residence time of the water body.
Hence, in this study we investigated gross (GNP), net (NNP) and substrate-induced nitrification potentials (SNP) and abundances of archaeal and bacterial amoA genes in the water column and in the sediment along the salinity gradient of the hyper-turbid, well-mixed Ems estuary in northwest of Germany, which is one of the German tributaries to the North Sea. Our key questions were (1) do nitrifications potentials vary across the salinity gradient (2) do water column and sediments are different with respect to the different nitrification potentials (3) does the ratio between archaeal and bacterial amoA gene abundances change across the salinity gradient and (4) which variables can be identified as controlling factors for the process of nitrification in water column and sediment?
Materials and methods
Study site
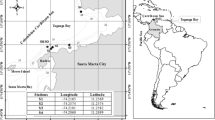
The Ems estuary is situated on the border between Germany and the Netherlands and discharges its water into the Wadden Sea in the southeastern part of the North Sea (Fig. 1). The tidal part of the River Ems, which is the main river draining into the Ems estuary is about 53 km long from the weir at Herbrum to Emden (Winterwerp et al. 2013). The River Ems serves as a draining canal and shipping route, and its embankments offer space for habitation and agriculture (Krebs and Weilbeer 2008). The catchment area of the Ems is 17,934 km2. The average discharge is 80.8 m3 s−1 (Krebs and Weilbeer 2008) and a residence time of 15–70 days depending on discharge (Middelburg et al. 2002). The Ems estuary is defined as a hyper-turbid estuary and the concentration of suspended sediment particles is high in the lower part of the river with up to 30–40 g dw L−1 (Talke et al. 2009). Deepening of the lower River Ems has strongly amplified the tides and increased the concentrations of suspended sediment within the tidal river (de Jonge et al. 2014). Bos et al. (2012) identifies the Ems estuary as a degraded ecological system, mainly because of a strongly artificial morphology, high level of turbidity, extended periods of oxygen minimum or anoxia situations in certain zones and a limited quality and quantity of estuarine habitats for flora and fauna. The nutrient loads are high (Gade et al. 2011). For our study, we have divided the Ems Estuary in three parts, the outer estuary, the Dollard Bay and the River Ems itself, which are polyhaline, mesohaline and oligohaline, respectively (Fig. 1). The River Ems has a depth of up to 5 m, while the water in the Dollard and in the outer Estuary was up to 20 m deep (Supplementary Table S1). The river and estuary kilometers of the Ems, as presented in Fig. 1, started in the town of Papenburg where two tributaries merge.
Map of the Ems estuary presenting the sampling stations. Blues dots represent the official River Ems kilometers started in Papenburg and the following offshore kilometer based on distance calculation by GIS, respectively. The sampling stations were marked by triangles: Black triangles indicate stations where only environmental parameters in the water column were measured, while red triangles present stations where additional water and sediment samples were taken for nitrification rate measurements, and for determining copy numbers of archaeal and bacterial amoA genes. (Color figure online)
Sampling
The sampling campaign was conducted with the R/V Ludwig Prandtl in August 2014. Water was collected with the use of an on-board membrane pump, and turbidity, salinity, fluorescence, pH and oxygen were measured continuously with the on-line in-situ Ferrybox system (Petersen et al. 2011), which has a resolution of one data point per minute. However, we present only the averages measured at the sampling stations in this paper. Water or water plus sediment samples were collected at distinct sampling stations, which are indicated in Fig. 1 by red and black triangles, respectively. Up- and downstream sampling cruises were conducted on three consecutive days (Figure S1). Samples were collected at the stations irrespective of the state of tide with the consequence of shifts in tide and current velocity between the sampling stations. Sediment samples were collected with a box corer. A mixed sample from the top 2–5 cm of the sediment was obtained from different box cores taken at the same location. At the Stations 280 and 279 in the oligohaline part of the estuary, the sediment samples were dark and muddy, indicating mainly anoxic conditions, while at the other stations more oxic, sandy sediments were found. The mixed sediment sample was used for nitrification potentials measurements, density and porosity determinations and DNA isolation. The sediment samples intended for DNA isolation were frozen and maintained at − 20 °C until extraction.
Water samples were either taken by a Niskin-Bottle 2 m above the bottom at the red-colored sampling stations (Fig. 1) or simply by a bypass of the Ferrybox system 2 m below the water surface at the black-colored stations. Water samples for nutrient and isotope analyses were immediately filtered through combusted (4 h, 450 °C) GF/F filters and stored in acid-washed (10% HCl, overnight) PE bottles. The filtered water samples were frozen on board and stored at − 20 °C until analyses started. Filtered samples for suspended matter analysis were dried at 50 °C and were also stored at − 20 °C. For DNA isolation from the water column the samples were filtered through a polycarbonate filter (0.2 µm) and the filters were kept at − 20 °C until DNA isolation started. The volume of the filtered water varied between 20 and 160 mL depending on the suspended matter (SPM) concentration.
Nutrient and sediment analysis
Duplicates of filtered water samples were analyzed for concentrations of ammonium, nitrite and nitrate, using an automated continuous flow system (AA3, Seal Analytical, Germany) and standard colorimetric techniques (Hansen and Koroleff 2007). In subsamples from nitrification potential rate measurements, ammonium was analyzed manually based on the same colorimetric techniques. Nitrite and nitrate concentrations in these subsamples were analyzed by HPLC (Meincke et al. 1992). Methods for density, porosity and water content measurements were based on Gonsiorczyk et al. (1995).
Nitrification potentials
For the assessment of nitrification potentials in the Ems estuary, we used two incubation approaches in microcosms, one based on isotope dilution and one based on substrate addition. The incubations were done separately for water and sediment samples. A long-term incubation for up to 15 days was used to encompass the lag-period and the period of linear increase of nitrate. In consequence, the incubations present nitrification potentials, because also microbial growth is included as well as nitrification-coupled processes like re-mineralization as a source of ammonium and denitrification as a sink of nitrate. Hence, the measured nitrification potentials are not comparable with in-situ rates, but likely overestimate these rates. The incubation time of 15 days is equivalent to the minimum residence time of water in the Ems estuary.
To measure gross and net nitrification potentials (GNP and NNP, respectively) in water samples, 120 mL of fresh water plus 50 µM end concentration of 15N-labelled NO3− were incubated. The incubation of the sediment occurred with 40 g fresh sediment plus 100 ml bottom water collected at the same location, to which 250 µM end concentration of 15N-labelled NO3− was added. This is a modified isotope dilution method (Inselsbacher et al. 2007). Both incubations were done on a rotary shaker (110 rpm) in 250 mL glass bottles with open caps in the dark at room temperature for 15 days. Sub-samples were taken every 2 or 3 days, centrifuged (15 min, 13,000 g), and nitrite and nitrate concentrations were determined immediately by HPLC. Incubations were continued until and nitrate concentrations in the samples were stable, but not longer than 15 days. In total, we conducted triplicate measurements for the water as well for the sediment incubations. The calculation of GNP was based on the 15N-nitrate dilution method of Norton and Stark (2011) using the changes in concentration and in δ15NNO3 values. For GNP the changes of δ15N in nitrate were included in the calculations (see Eq. 1); the steepest, linear part of the slope corresponds with the nitrification rate.
where P is the nitrate concentration, I is the 15 N atom % excess, t is the length of incubation time and 0 and t indicate the time points. The NNP was calculated by plotting the change in nitrite and nitrate concentration versus time (typical raw data presented in Figure S2). In the 15N-labeled incubations we analyzed the isotopic composition of nitrate using the denitrifier method (Casciotti et al. 2002; Sigman et al. 2001), which is based on the isotopic analysis of nitrous oxide (N2O) produced from nitrate and nitrite by the denitrifying bacterium Pseudomonas aureofaciens (ATCC#13985). This method is descripted in detail elsewhere (Dähnke et al. 2008).
For the determination of SNP, 30 g fresh sediment and 75 mL of bottom water (Stations 276–278) or 40 g fresh sediment and 100 mL bottom water (Stations 279 and 280) enriched with 0.5 mM ammonium sulfate were incubated for 15 days in the dark at 20 °C on a rotary shaker (100 rpm). Sub-samples were taken daily. SNP were calculated by the increase of nitrite plus nitrate versus time measured during the linear phase according to the protocol of Belser and Mays (Belser and Mays 1980), as modified by Verhagen and Laanbroek (1991), which occurred mainly in the first 5 days of incubation (typical raw data presented in Figure S2).
DNA extraction and quantitative PCR assay
DNA was extracted from water as well as from sediment samples by the MoBio PowerSoilKit (MoBio, CA, USA) according to the manufacturer’s instructions. For the water column samples, filtered material on polycarbonate filters was used for the extraction of DNA. Duplicates were obtained by cutting each filter in two. For the sediment samples, duplicate 1 g sediment samples were extracted. The DNA samples were stored at − 20 °C until further processed. The DNA concentrations ranged from 0.4 to 15.2 ng µL−1 as measured by a Nanodrop spectrophotometer [Nanodrop Technology, Wilmington, DE (Delaware), USA]. The copy numbers of archaeal and bacterial amoA genes were quantified using the primer set Arch-amoAF and Arch-amoAR (Francis et al. 2005), and amoA-1F and amoA-2R (Rotthauwe et al. 1997), respectively. The bacterial primer amplified just the amoA gene of the Betaproteobacteria and excludes the Gammaproteobacteria (Rotthauwe et al. 1997). Each reaction was performed in a 20 µL volume containing 5 µL DNA solution, 1 µL of a 5 µM solution of each primer, 1 µL BSA and 10 µL of SYBR Green thereby using the Rotor-Gene SYBER Green PCR Kit (Qiagen, Hilden, Germany), specific for the Rotor-Gene 6000 (Qiagen, Hilden, Germany). In the quantitative PCR (qPCR) assay, clones of E. coli JM109 containing the archaeal or the bacterial amoA gene fragment were included as standard, generating standard curves with up to seven dilutions. A control reaction without template DNA was included in each qPCR assay. All DNA samples and the negative control were analyzed in duplicates to obtain an accurate value for the amoA gene abundance in each sample. The standard derivation in one run was never larger than 1.5%. The qPCR was conducted in 40 cycles, the annealing temperature was 56 and 55 °C for the archaeal and bacterial amoA genes, respectively. Amplification efficiencies of 95–110% were obtained for the archaeal and bacterial amoA, respectively, with R2 values of 0.9988 and 0.9982 and with slopes of − 3.04 and − 3.51.
Statistical analysis
All statistical analyses including Principle Component analysis, the test for normal distribution, the one-way ANOVA with Tukey’s HSD Post Hoc test for differences between means across groups, the non-parametric Kruskal–Wallis test for comparing distributions across groups, the non-parametric Wilcoxon test for comparing median differences between relating samples, and Spearman rank correlation analysis, were performed with the IBM-SPSS software package version 23 (IBM Corp. Armonk, NY).
Results
Water and sediment properties
Water properties varied between the different sampling stations (Supplementary Table S1). Water salinity ranged from 0.4 to 31.5 PSU in a gradient from the River Ems through the Dollard into the Wadden Sea (Fig. 2a). The oxygen saturation increased with increasing salinity (Fig. 2b). The lowest oxygen saturation values were found in the River Ems where salinities were below 3 PSU. The oxygen concentration in this oligohaline part of the estuary ranged from 40 to 54 µmol L−1, while oxygen concentrations below 62.5 µmol L−1 are assumed to represent hypoxic conditions (Friedrich et al. 2014). The fluorescence (Fig. 2c) of the water column, being an indicator for the presence of phytoplankton, was relatively low and decreased with increasing salinity. The turbidity decreased strongly with increasing salinity, especially in the river and Dollard area. Similarly, the suspended matter concentration changed, but there was no significant correlation between SPM and turbidity (Fig. 2d, e), which could be due to the use of different methods, i.e. weighting filters versus using a sensor, respectively. The amount of suspended matter at sampling Station 280 at km point 22 (i.e. 18.5 g L−1) was above the mean amount of 31 mg L−1 measured at the other stations. Station 280 represented the high turbidity zone of the estuary. With a water temperature of 24.8 °C (Supplementary Table S1), sampling Station 276 was above the mean water temperature of 23.0 °C. This increased temperature may represent water mass distinct from the water masses sampled at the other stations. Since both water temperature and amount of suspended matter are likely controlling factors with respect to nitrification rates, the outliers were retained in further statistical analyses.
Water column characteristics along the River Ems estuary. a Salinity; b oxygen saturation; c fluorescence (estimation for chlorophyll); d turbidity; e suspended matter (SPM). The red triangles represent the Stations 280, 279, 278, 277, and 276, respectively, where additional water and sediment samples were collected for rates and molecular measurements. (Color figure online)
Leaving the Dollard, ammonium as well as nitrite increased with increasing salinity up to 8.5 and 3.5 µmol L−1, respectively (Fig. 3a, b). The major form of dissolved inorganic nitrogen (DIN) was nitrate, which clearly decreased with increasing salinity (Fig. 3c). Nitrate was highest at low salinity (177 µmol L−1) and decreased (4 µmol L−1) into the estuary and Wadden Sea.
Dissolved inorganic nitrogen concentrations along the River Ems estuary. a Ammonium; b nitrite; c nitrate. The red triangles represent the Stations 280, 279, 278, 277, and 276, respectively, where additional water and sediment samples were collected for rate and molecular measurements. (Color figure online)
Both density and porosity of the sediment increased at sites with increasing salinity from the river into the outer estuary, especially at the oligohaline Stations 278–280 in the River Ems itself; the porosity was low with 0.70–0.52. The water content of the sediment decreased with increasing salinity (Supplementary Table S1).
A principal component analysis on the environmental factors measured in the water column of all stations yielded three principal components that explained 92.7% of the variance observed with these factors. Salinity, pH, oxygen, ammonium and nitrite contributed largely and positively to the first principal component (PC1) that explained 73.2% of the total variance. Turbidity, fluorescence and nitrate contributed also to this principle component, but in a negative way. Temperature contributed mostly and positively to the second principle component (PC2) that explained 10.2% of the total variance, and the amount of suspended matter contributed mostly and positively to the third principle component (PC3), which explained 9.3% of the total variance (Supplementary Table S4).
Nitrification potentials in water column and sediment
Nitrification potentials were measured in water as well as in sediment samples collected at five distinct sampling stations along the salinity gradient of the Ems estuary (Fig. 1, typical raw data presented in Figure S2). Whereas GNP and NNP were measured in water column and in sediment, SNP was only determined in sediments. In the water column, GNP and NNP ranged from 0.7 to 6.7 µmol L−1 day−1 and were highest at the oligohaline Station 280 at km point 22 and decreased with increasing salinity until a minimum was reached at the mesohaline Station 279 at km point 35 (Fig. 4a). Downstream from this station, nitrification rates increased again at the oligohaline Station 277 and 276 at km points 52 and 63, respectively. GNP but not NNP were normally distributed according to the Shapiro–Wilk test. The distributions of the means of GNP and NNP were significantly (p = 0.002) different according to the Wilcoxon test and GNP and NNP were significantly (p < 0.001) correlated (Supplementary Table S2).
Nitrification rates (a), archaeal and bacterial amoA gene copy numbers (b) and ratios of ammonia-oxidizing archaea and bacteria (c) measured in the water column of the Ems estuary. NNP Net nitrification potential, GNP gross nitrification potential. AOA Ammonia-oxidizing archaea and AOB ammonia-oxidizing bacteria
In the sediments, the highest GNP and NNP were observed at the mesohaline Station 278 at km point 35 with approx. 110 nmol N gdw−1 day−1 (Fig. 5a). Due to decreasing nitrate concentrations during the incubations, no NNP could be calculated at the oligohaline Stations 280 and 279 at km points 22 and 26. The SNP exceeded the gross nitrification rates at the oligohaline Stations 280 and 279 at km points 22 and 26 by a factor of four. At the downstream mesohaline and polyhaline Stations 278, 277 and 276 at km points 35, 52 and 63, respectively, the SNP, GNP and NNP were rather similar at approximately 20 nmol N gdw−1 day−1.
Nitrification rates (a), archaeal and bacterial amoA gene copy numbers (b) and ratios of ammonia-oxidizing archaea and bacteria (c) measured in the sediment of the Ems estuary. NNP Net nitrification potential, GNP gross nitrification potentials, SNP substrate-induced nitrification potential, AOA ammonia-oxidizing archaea and AOB ammonia-oxidizing bacteria
SNP, GNP and NNP in the sediment were not normally distributed according to the Shapiro–Wilk test. The distributions of the means of SNP, GNP and NNP in the sediment were significantly different according to the Wilcoxon test (p = 0.017 for net versus gross, p = 0.001 for net versus potential, and p = 0.013 for gross versus potential). In the sediment, no significant correlations were observed between the different nitrification rates in the sediment (Supplementary Table S2).
Abundances of archaeal and bacterial amoA genes in water and sediment
Archaeal amoA genes outnumbered the bacterial amoA genes by two orders of magnitude both in the water column and in the sediment, while the gene abundances generally decreased with increasing salinity, especially in the sediment (Figs. 4b, 5b). In the water column samples, the gene numbers of archaeal amoA ranged from 5.03 × 104 to 2.26 × 106 gene copies L−1 (Fig. 4b). The highest gene copy numbers were found at the oligohaline Station 279 at km point 26 where the lowest oxygen saturation and the highest turbidity and suspended matter concentrations were measured. The gene copy numbers of bacterial amoA were low and hardly detectable in the water column (detection limit ~ 103 L−1). But once again the highest gene abundance was found at Station 279 with 2.26 × 104 genes copies L−1 (Fig. 4b). The bacterial gene abundance in the water column was low at Station 276. Archaeal and bacterial amoA genes were not detectable at sampling Station 277 at km point 52.
The gene abundance of archaeal amoA in the sediment varied from 6.6 × 104 to 2.5 × 109 copies g dw−1 (Fig. 5b). The highest gene abundance was observed in samples from sampling Stations 280 and 278 at km points 22 and 35, respectively, while the gene copy numbers at Station 279 at km point 26 were lower. The gene abundances of bacterial amoA were lower and ranged between 2.4 × 104 in the outer estuary to 3.1 × 107 copies g dw−1 at the mesohaline Station 278 at km 35 (Fig. 5b). In the polyhaline stations the gene abundance was lower than in the oligohaline or mesohaline stations.
In the water column as well as in the sediment, the log numbers of archaeal and bacterial amoA gene copies, as well as the log of the ratio between them, were not normally distributed. Only in the sediment, the distributions of the medians of archaeal and bacteria amoA gene copy numbers were significantly (p = 0.043) different according to the Wilcoxon test.
Discussion
Nitrification potentials in the Ems estuary
Although it is evident that microbial processes both in the water column and sediment control the nature of nitrogen in estuaries, nearly all studies related to the process of nitrification in river mouths focused either on sediments or on the water column. A recent paper of Damashek et al. (2016) listed published nitrification rates from the water columns of different estuaries. The reported nitrification rates in estuaries are mainly higher than in marine waters, but can vary across a broad range from zero to more than 360 µmol L−1day−1. This variation in rates is probably due to differences in salinity, amount of suspended matter and ammonium concentration (Damashek et al. 2016). Unfortunately, the rates are difficult to compare because different approaches have been used. Normally short-term incubations of 6–48 h were applied. Nevertheless, the potentials of 0.7–6.7 µmol L−1 day−1 measured in the water column of the Ems estuary fall within the range listed by Damashek et al. (2016), but were determined in long-term incubations. As mentioned already in the Methods section, the measured potentials will likely overestimate the nitrification in the water column, because the incubations include microbial growth and also nitrification coupled process of mineralization. A direct comparison to rates measured in the Elbe estuary was possible because of using the same method. With up to 7 µmol L−1 day−1, nitrification rates in the port of Hamburg situated in the Elbe estuary, are slightly higher than those observed in the Ems estuary (Sanders et al. 2017). Like the Ems estuary, the Elbe estuary showed higher rates in the freshwater part.
By measuring GNP and NNP as well as SNP in the sediment samples, we got an estimation of the ranges of the nitrification potentials in the sediment when ammonium or oxygen is not limiting nitrification. The nitrification potentials measured in the sediments of the Ems estuary showed clearly that the substrate-induced potentials at the oligohaline sampling stations are higher than the gross and net potentials. At the mesohaline and polyhaline stations the SNP, GNP and NNP are more similar. In the oligohaline part the available ammonium seems to be a limiting factor, so that by ammonium addition the potentials rise. The main source for ammonium is the mineralization of organic matter as was observed before in the freshwater part of the Elbe estuary (Sanders et al. 2017). However, the organic matter in the oligohaline part seems to be refractory without much fresh material, because the C/N values of the suspended matter were higher than 10 (data not shown) and the fluorescence, an indicator for fresh phytoplankton, were low (Fig. 2). Further along the estuary, ammonium seems to be available from mineralization of fresh organic matter originating from the North Sea (Fig. 1). Despite the long incubation times, the measured SNP in the Ems estuary were lower than those observed in other estuarine sediments like in sediments from the Elkhorn Slough estuary and the Sacramento–San Joaquin Delta, both in California, which used shorter incubations times and which ranged from 5 to 4608 nmol g−1 day−1 and from 192 to 3504 nmol g−1 day−1, respectively (Damashek et al. 2015; Smith et al. 2015).
One other reason for the lower SNP in the sediments of the Ems estuary, at least in the oligohaline sediments, could be the hypoxic conditions in the water column. Such hypoxic conditions might also have been the cause of the absence of net nitrification in sediments located in that part of the estuary. Nitrate produced during the process of gross nitrification was apparently directly consumed by nitrate-consuming processes, of which nitrate reduction by denitrification is a likely candidate. Nitrate immobilization is probably of less importance due to low numbers of phototrophic algae as can be inferred from the low fluorescence values measured in the water column. The non-conservative mixing behavior of nitrate in the Ems estuary (Fig. 6) indicates a nitrate loss in the salinity gradient between 0 and 22 PSU, whereas in the outer estuary a nitrate gain is apparent. Middelburg and Nieuwenhuize (2000) presented data from the late 1990s and found that there was a nitrate loss at lower salinities of turbid, tidal estuaries. Increased amounts of ammonium, caused by a net input from extensive tidal-flat areas, are consecutively nitrified and resulted in a net nitrate gain at higher salinities.
Ammonia oxidizer abundances
From the ratio between archaeal and bacterial amoA gene copy numbers, it can be concluded that the ammonia-oxidizing archaea dominated the oligohaline and mesohaline sampling Stations 278–280, both in the sediment and in the water column, whereas more comparable archaeal and bacterial amoA gene abundances were observed at the polyhaline sampling stations. The ratio between archaeal and bacterial amoA gene copy numbers decreased with increasing salinity both in the water column and in the sediment. A decline in this ratio with increasing salinities has also been observed at Huntington Beach, California (Santoro et al. 2008) and at Plum Island Sound, Massachusetts (Bernhard et al. 2010). In contrast, an increase in the ratio of archaeal and bacterial amoA gene copy numbers was observed in the hyper-eutrophic Yong River in China (Zhang et al. 2015). Hence, in addition to salinity other environmental factors must determine the ratio between the copy numbers of the archaeal and bacterial amoA genes. In a study comprising sediments from six different estuaries, Caffrey et al. (2007) observed a dominance of ammonia-oxidizing archaea, except for one high ammonium estuary where ammonia-oxidizing bacteria dominated. In a tidal marsh in south-eastern Connecticut, Moin et al. (2009) detected always a dominance of archaeal over bacterial amoA gene copies. In contrast, ammonia-oxidizing bacteria dominated in the sediments of Elkhorn Slough, a small agriculture-impacted coastal estuary that opens into Monterey Bay in California (Wankel et al. 2011). In sediments at the landward reaches of the San Francisco Bay, Damashek et al. (2015) observed strong regional differences in ammonia-oxidizing communities with ammonia-oxidizing bacteria dominating in ammonium-rich sediments. In freshwater systems a similar heterogeneous assembly of archaeal and bacterial ammonia oxidizers was described, but ammonia-oxidizing bacteria were again dominant in ammonium-rich creeks (Herrmann et al. 2011). Environmental constraints, including most likely ammonium availability, seem to govern the dominance of either one of the ammonia-oxidizing microbial groups.
Comparison between water column and sediment
To compare the absolute impact of water column and sediments on the nitrification characteristics of the hyper-turbid Ems estuary, we converted the GNP and the amoA gene numbers, which were both expressed per g dry sediment, in comparable units per L of sediment by application of the bulk density determined from each station. Assuming that the upper 5 cm of the sediment contains the potentially active ammonia-oxidizing cells, the GNP and the amoA gene numbers below 1 m2 of sediment can be calculated. These rates and numbers can then be compared with the rates and gene numbers above 1 m2 of water volume taking into account the local depth of the water column at the same sampling station. The ratio of these rates and amoA gene numbers per m2 reflect the relative impact of water column and sediment on the process of nitrification. With increasing water depth the relative impact of the water column nitrification increased clearly (Fig. 7a, b).
Ratios of archaeal and bacterial amoA gene abundance and gross nitrification rates. a amoA gene archaea and bacteria observed in water column versus sediment of the Ems estuary converted to copies per m2. b Ratio of gross nitrification rates obverted in water column versus sediment converted to µmol per dm2 and day
Remarkably, the water to sediment ratios of both archaeal and bacterial amoA gene copy numbers were significantly correlated (p < 0.001, Supplementary Table S2) as can also be observed in Fig. 7a. This correlation suggests that both variables are governed by the same factor, which is likely the resuspension of sediment particles with their attached ammonia-oxidizing microorganisms in the water column. Although we have not determined particle-associated nitrification rates in the water column of the Ems estuary, the significant and positive correlation between both GNP and NNP, and the third principle component of a PCA analysis that largely represents the amount of suspended particulate matter, makes it likely that a large part of the active ammonia-oxidizing cells was particle-associated as well. In the tidal River Elbe, the contribution of attached bacteria to the total number ranged from 50 to 100% depending on the load with suspended particulate matter in the water (Stehr et al. 1995). In the Scheldt estuary particle-associated nitrification rates varied between 57 and 80% depending on the month of sampling (de Bie et al. 2002). The local similarity in water—sediment partitioning of archaeal and bacterial amoA genes along the estuary of the Ems River suggests a close relationship between the ammonia-oxidizing communities in sediment and water column. The existence of such a close relationship would be strengthened by an analysis of the species diversity of ammonia-oxidizing archaea and bacteria in sediment and water column. The hyper-turbid Ems estuary seems to be a well-suited environment to conduct such analyses.
Environmental factors and potential controlling factors affecting nitrification
Some significant correlations were found in a Spearman rank correlation analysis between the three principle components and the nitrification-related factors measured in the water column (Table 1). The GNP and NNP in the water column correlated significantly (p = 0.037) and positively with the third principle component, i.e. with the amount of suspended particulate matter, which could imply that nitrification in the water column was largely particle-bound. Also Damashek et al. (2016) observed a significant correlation between nitrification rates and the amounts of suspended particulate matter in turbid, nutrient-rich estuarine waters of San Francisco Bay. Also in the water column of the Ems estuary, the log of the archaeal amoA gene copy numbers, but not the log of the copy numbers of the bacterial amoA gene, correlated significantly (p < 0.001) and positively with the first principle component, which was mainly positively affected by salinity, oxygen, ammonium and nitrite and negatively by turbidity, fluorescence and nitrate. Hence, nitrification rates and abundance of archaeal amoA genes were governed by different environmental factors, which agree with the absence of a significant correlation between potentials and numbers of amoA gene copies. So, it may cautiously be concluded that a large fraction of the amoA gene pool in the water column of the Ems estuary is not actively involved in the process of ammonia oxidation. This agrees with the conclusion of Bernhard and Bollmann (2010), that there is not necessarily a causal relationship between rates and gene abundance. An explanation could be that ammonia-oxidizing archaea can also grow mixotrophic and use other energy sources (Qin et al. 2014) or that we overlooked the ammonia-oxidizing Gammaproteobacteria, because of the primer set used. For identifying active parts of the ammonia-oxidizing community, the analysis of amoA gene transcripts may be a solution. However it should be kept in mind that especially the ammonia-oxidizing archaea may be able to maintain a relatively high level of amoA gene transcripts during the stationary phase as was shown for archaeal oxidizers enriched from freshwater (French and Bollmann 2015).
A Spearman rank correlation analysis between measured sediment characteristics and nitrification-coupled factors showed a significant (p = 0.037) correlation between the log of the copy numbers of the archaeal amoA gene on one side and the water content of the sediment (positive) and the porosity (negative) on the other side (Supplementary Table S3). So the higher the water content and the lower the porosity of the sediment, the higher the abundance of the archaeal amoA genes. However, this does not necessarily mean that water content and porosity are controlling the abundance of archaeal amoA genes as both factors are part of a complex of sediment characteristics connected to the estuarine gradient that had not been determined. The log of the bacterial amoA gene copy numbers did not show a significant correlation with one of the sediment characteristics.
Concluding remarks
To underpin the importance of the process of nitrification for the Ems estuary, a follow-up study on the coupling of this process with other nitrogen-converting processes should be done. For example, a question that remains from this present study is the existence of a direct coupling between nitrification and nitrate reduction in the top 10 cm layers of the sediment leading to zero or low NNP as measured in the oligohaline and polyhaline sediments, respectively. Why was nitrate consumption lacking in the mesohaline sediments of the Dollard, where GNP and NNP were rather similar? The coupling of other N-cycle processes like denitrification and mineralization to nitrification will change the filter function of the estuary to a sink or a source of nitrate, respectively. The study of the N-cycle in estuaries will help to understand the impact of estuaries on the eutrophication of coastal waters and to learn how that can be changed.
References
Belser L, Mays E (1980) Specific inhibition of nitrite oxidation by chlorate and its use in assessing nitrification in soils and sediments. Appl Environ Microbiol 39:505–510
Bernhard AE, Bollmann A (2010) Estuarine nitrifiers: New players, patterns and processes Estruarine. Coast Shelf Sci 88:1–11
Bernhard AE, Colbert D, McManus J, Field KG (2005a) Microbial community dynamics based on 16S rRNA gene profiles in a Pacific Northwest estuary and its tributaries. FEMS Microbiol Ecol 52:115–128
Bernhard AE, Donn T, Giblin AE, Stahl DA (2005b) Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ Microbiol 7:1289–1297. https://doi.org/10.1111/j.1462-2920.2005.00808.x
Bernhard AE, Tucker J, Giblin AE, Stahl DA (2007) Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ Microbiol 9:1439–1447. https://doi.org/10.1111/j.1462-2920.2007.01260.x
Bernhard AE, Landry ZC, Blevins A, de la Torre JR, Giblin AE, Stahl DA (2010) Abundance of ammonia-oxidizing archaea and bacteria along an Estuarine salinity gradient in relation to potential nitrification rates. Appl Environ Microbiol 76:1285–1289. https://doi.org/10.1128/aem.02018-09
Bollmann A, Laanbroek HJ (2002) Influence of oxygen partial pressure and salinity on the community composition of ammonia-oxidizing bacteria in the Schelde estuary. Aquat Microb Ecol 28:239–247. https://doi.org/10.3354/ame028239
Bos D, Büttger H, Esselink P, Jager Z, De Jonge V, Van Maren B, Schuchardt B (2012) The ecological state of the Ems estuary and options for restoration. A&W Report
Caffrey JM, Bano N, Kalanetra K, Hollibaugh JT (2007) Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J 1:660–662. https://doi.org/10.1038/ismej.2007.79
Casciotti KL, Sigman DM, Hastings MG, Bohlke JK, Hilkert A (2002) Measurement of the oxygen isotopic composition of nitrate in seawater and freshwater using the denitrifier method. Anal Chem 74:4905–4912
Crossland CJ et al (2005) The coastal zone—a domain of global interactions. In: Crossland CJ, Kremer HH, Lindeboom HJ, Marshall Crossland JI, Le Tissier MDA (eds) Coastal fluxes in the anthropocene: the land–ocean interactions in the coastal zone project of the international geosphere–biosphere programme. Springer, Berlin, pp 1–37. https://doi.org/10.1007/3-540-27851-6_1
Dähnke K, Bahlmann E, Emeis K (2008) A nitrate sink in estuaries? A assessment by means of stable nitrate isotopes in the Elbe estuary limnol. Oceanography 53:1504–1511
Daims H et al (2015) Complete nitrification by Nitrospira bacteria. Nature 528:504–509
Damashek J, Smith JM, Mosier AC, Francis CA (2015) Benthic ammonia oxidizers differ in community structure and biogeochemical potential across a riverine delta. Front Microbiol. https://doi.org/10.3389/fmicb.2014.00743
Damashek J, Casciotti KL, Francis CA (2016) Variable nitrification rates across environmental gradients in turbid, nutrient-rich Estuary waters of San Francisco Bay. Estuaries Coasts 39:1050–1071. https://doi.org/10.1007/s12237-016-0071-7
de Bie MJM et al (2001) Shifts in the dominant populations of ammonia-oxidizing beta-subclass proteobacteria along the eutrophic Schelde estuary. Aquat Microb Ecol 23:225–236
de Bie MJM, Starink M, Boschker HTS, Peene JJ, Laanbroek HJ (2002) Nitrification in the Schelde estuary: methodological aspects and factors influencing its activity. FEMS Microbiol Ecol 42:99–107. https://doi.org/10.1111/j.1574-6941.2002.tb00999.x
de Jonge VN, Schuttelaars HM, van Beusekom JEE, Talke SA, de Swart HE (2014) The influence of channel deepening on estuarine turbidity levels and dynamics, as exemplified by the Ems estuary Estuarine. Coast Shelf Sci 139:46–59. https://doi.org/10.1016/j.ecss.2013.12.030
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns sediments of the ocean. PNAS 102:14683–14688. https://doi.org/10.1073/pnas.0506625102
French E, Bollmann A (2015) Freshwater ammonia-oxidizing archaea retain amoA mRNA and 16S rRNA during ammonia starvation. Life 5:1396–1404
Friedrich J et al (2014) Investigating hypoxia in aquatic environments: diverse approaches to addressing a complex phenomenon. Biogeosciences 11:1215–1259. https://doi.org/10.5194/bg-11-1215-2014
Gade R, Fischer M, Kaiser R, Rabe O, Grage A, Knaack J, Petri G, Trepel M, Brockmann U, von Beusekom J, Kuhn U (2011) Konzept zur Ableitung von Nährstoffreduzierungszielen in den Flussgebieten Ems, Weser, Elbe und Eider aufgrund von Anforderungen an den ökologischen Zustand der Küstengewässer gemäß Wasserrahmenrichtlinie. Ad-hoc-AG Nährstoffreduzierung des BLMP, Bund Länder, Messprogramm / Umsetzung der Wasserrahmenrichtlinie, 50 S
Galloway JN, Aber JD, Erisman JW, Seitzinger SP, Howarth RW, Cowling EB, Cosby BJ (2003) The nitrogen cascade. BioScience 53:341–356. https://doi.org/10.1641/0006-3568(2003)053[0341:tnc]2.0.co;2
Gonsiorczyk T, Casper P, Koschel R (1995) Vergleichende Sedimentuntersuchungen zum Kohlenstoff- und Phosphorgehalt in Baltischen Seen. Limnol Ecol Manag Inland Waters 25:365–379
Hansen HP, Koroleff F (2007) Determination of nutrients. Methods Seaw Anal. https://doi.org/10.1002/9783527613984.ch10
Heiss EM, Fulweiler RW (2016) Coastal water column ammonium and nitrite oxidation are decoupled in summer Estuarine. Coast Shelf Sci 178:110–119. https://doi.org/10.1016/j.ecss.2016.06.002
Herrmann M, Scheibe A, Avrahami S, Kusel K (2011) Ammonium availability affects the ratio of ammonia-oxidizing bacteria to ammonia-oxidizing archaea in simulated creek ecosystems. Appl Environ Microbiol 77:1896–1899. https://doi.org/10.1128/aem.02879-10
Howarth RW, Marino R (2006) Nitrogen as the limiting nutrient for eutrophication in coastal marine ecosystems: evolving views over three decades. Limnol Oceanogr 51:364–376. https://doi.org/10.4319/lo.2006.51.1_part_2.0364
Inselsbacher E, Cambui CA, Richter A, Stange CF, Mercier H, Wanek W (2007) Microbial activities and foliar uptake of nitrogen in the epiphytic bromeliad Vriesea gigantea. New Phytol 175:311–320. https://doi.org/10.1111/j.1469-8137.2007.02098.x
Krebs M, Weilbeer H (2008) Ems-dollart estuary. Die Küste 74:252–262
Meincke M, Bock E, Kastrau D, Kroneck PMH (1992) Nitrite oxidoreductase from nitrobacter hamburgensis: redox center and their catalytic role. Arch Microbiol 158:127–131
Middelburg JJ, Nieuwenhuize J (2000) Uptake of dissolved inorganic nitrogen in turbid, tidal estuaries. Mar Ecol Prog Ser 192:79–88
Middelburg J, Nieuwenhuize J (2001) Nitrogen isotope tracing of dissolved inorganic nitrogen behaviour in tidal estuaries Estuarine. Coast Shelf Sci 53:385–391
Middelburg JJ et al (2002) Methane distribution in European tidal estuaries. Biogeochemistry 59:95–119. https://doi.org/10.1023/a:1015515130419
Moin NS, Nelson KA, Bush A, Bernhard AE (2009) Distribution and diversity of archaeal and bacterial ammonia oxidizers in salt marsh sediments. Appl Environ Microbiol 75:7461–7468. https://doi.org/10.1128/aem.01001-09
Mosier AC, Francis CA (2008) Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol 10:3002–3016. https://doi.org/10.1111/j.1462-2920.2008.01764.x
Norton JM, Stark JM (2011) Regulation and measurement of nitrification in terrestrial systems. In: Klotz MG (ed) Methods in enzymology, vol 486. Elsevier, Amsterdam, pp 343–368
Pätsch J, Serna A, Dähnke K, Schlarbaum T, Johannsen A, Emeis K-C (2010) Nitrogen cycling in the German Bight (SE North Sea)—clues from modelling stable nitrogen isotopes. Cont Shelf Res 30:203–213. https://doi.org/10.1016/j.csr.2009.11.003
Petersen W, Schroeder F, Bockelmann F-D (2011) FerryBox—application of continuous water quality observations along transects in the North Sea. Ocean Dyn 61:1541–1554. https://doi.org/10.1007/s10236-011-0445-0
Qin W et al (2014) Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci 111:12504–12509. https://doi.org/10.1073/pnas.1324115111
Radach G, Pätsch J (2007) Variability of continental riverine freshwater and nutrient inputs into the North Sea for the years 1977–2000 and its consequences for the assessment of eutrophication. Estuaries Coasts 30:66–81. https://doi.org/10.1007/BF02782968
Rotthauwe J-H, Witzel K-P, Liesack W (1997) The Ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Sahan E, Muyzer G (2008) Diversity and spatio-temporal distribution of ammonia-oxidizing archaea and bacteria in sediments of the Westerschelde estuary. FEMS Microbiol Ecol 64:175–186. https://doi.org/10.1111/j.1574-6941.2008.00462.x
Sanders T, Schöl A, Dähnke K (2017) Hot spots of nitrification in the Elbe estuary and their impact on nitrate regeneration. Estuaries Coasts. https://doi.org/10.1007/s12237-017-0264-8
Santoro AE, Francis CA, De Sieyes NR, Boehm AB (2008) Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ Microbiol 10:1068–1079. https://doi.org/10.1111/j.1462-2920.2007.01547.x
Sigman DM, Casciotti KL, Andreani M, Barford C, Galanter M, Böhlke JK (2001) A bacterial method for the nitrogen isotopic analysis of nitrate in seawater and freshwater. Anal Chem 73:4145–4153. https://doi.org/10.1021/ac010088e
Smith JM, Mosier AC, Francis CA (2015) Spatiotemporal relationships between the abundance, distribution, and potential activities of ammonia-oxidizing and denitrifying microorganisms in intertidal sediments. Microb Ecol 69:13–24. https://doi.org/10.1007/s00248-014-0450-1
Stehr G, Böttcher B, Dittberner P, Rath G, Koops H-P (1995) The ammonia-oxidizing nitrifying population of the river Elbe estuary. FEMS Microbiol Ecol 17:177–186. https://doi.org/10.1111/j.1574-6941.1995.tb00141.x
Talke SA, de Swart HE, De Jonge V (2009) An idealized model and systematic process study of oxygen depletion in highly turbid estuaries. Estuaries Coasts 32:602–620
van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lücker S (2015) Complete nitrification by a single microorganism. Nature 528:555. https://doi.org/10.1038/nature16459
Verhagen FJM, Laanbroek HJ (1991) Competition for ammonium between nitrifying and heterotrophic bacteria in dual energy-limited chemostats. Appl Environ Microbiol 57:3255–3263
Wankel SD, Mosier AC, Hansel CM, Paytan A, Francis CA (2011) Spatial variability in nitrification rates and ammonia-oxidizing microbial communities in the agriculturally impacted Elkhorn Slough estuary California. Appl Environ Microbiol 77:269–280. https://doi.org/10.1128/aem.01318-10
Winterwerp JC, Wang ZB, van Braeckel A, van Holland G, Kösters F (2013) Man-induced regime shifts in small estuaries—II: a comparison of rivers. Ocean Dyn 63:1293–1306
Zhang Q, Tang F, Zhou Y, Xu J, Wang M, Laanbroek HJ (2015) Shifts in the pelagic ammonia-oxidizing microbial communities along the eutrophic estuary of Yong River in Ningbo City. China Front Microbiol. https://doi.org/10.3389/fmicb.2015.01180
Acknowledgements
We thank the crew of the R/V Ludwig Prandtl for cruise support and assisting with sampling. Markus Ankele helped us to analyse nutrients, and our colleagues at the department Operational Systems of the Helmholtz Zentrum Geesthacht provided the Ferrybox data. Roos Keijzer assisted with the qPCR analysis. This study was partly funded by the Helmholtz Association (VH-20 NG-721).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
27_2018_584_MOESM2_ESM.docx
Table S2: Results of a Spearman rank correlation analysis between ammonia oxidation-related characteristics determined in the water column and in the sediment of the Ems estuary. The correlation coefficients (Spearman’s rho) are presented above the diagonal, the two-tailed significant values below the diagonal. NNP = net nitrification potential, GNP = gross nitrification potential, SNP = substrate-induced nitrification potential, AOA = copy numbers of the archaeal amoA gene, and AOB = copy numbers of the bacterial amoA gene (DOCX 14 KB)
27_2018_584_MOESM3_ESM.docx
Table S3: Results of a Spearman rank correlation analysis between ammonia oxidation-coupled characteristics determined in the sediment of the Ems estuary. The correlation coefficients (Spearman’s rho) are presented above the diagonal, the two-tailed significant values below the diagonal. NNP = net nitrification potential, GNP = gross nitrification potential, SNP = substrate-induced nitrification potential, AOA = copy numbers of the archaeal amoA gene, and AOB = copy numbers of the bacterial amoA gene (DOCX 12 KB)
27_2018_584_MOESM4_ESM.docx
Table S4: List of initial Eigenvalues (A) and a Rotated Component matrix (rotation converged in 5 iterations) (B) of a Principal Component Analysis based on pelagic environmental factors with the exclusion of turbidity (DOCX 13 KB)
27_2018_584_MOESM5_ESM.tif
Figure S1: Effect of tide on the distribution of salinity in the Ems estuary. Sampling dates and upstream and downstream cruise measured in August 2014 are explained in the legend (TIF 3044 KB)
27_2018_584_MOESM6_ESM.tif
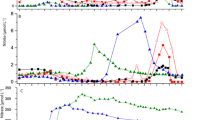
Figure S2: Incubation data nitrification rates measurements for Station 277 and 280. Incubation in water (A) for GNP and NNP, in sediments [B] for GNP and NNP and in sediments (C) for SNP. Nitrate and δ15NNitrate versus time [day]. The red colored sign in C indicate the period used for calculation of the rates (TIF 13377 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sanders, T., Laanbroek, H.J. The distribution of sediment and water column nitrification potential in the hyper-turbid Ems estuary. Aquat Sci 80, 33 (2018). https://doi.org/10.1007/s00027-018-0584-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00027-018-0584-1