Abstract
The oncoprotein c-Fos has been commonly found differently expressed in cancer cells. Our previous work showed that mouse cells lacking the immediate-early gene c-fos are hypersensitive to ultraviolet (UVC) light. Here, we demonstrate that in human diploid fibroblasts UV-triggered induction of c-Fos protein is a delayed and long-lasting event. Sustained upregulation of c-Fos goes along with transcriptional stimulation of the NER gene xpf, which harbors an AP-1 binding site in the promoter. Data gained on c-Fos knockdown and c-Fos overexpressing human cells provide evidence that c-Fos/AP-1 stimulates upregulation of XPF, thereby increasing the cellular repair capacity protecting from UVC-induced DNA damage. When these cells are pre-exposed to a low non-toxic UVC dose and challenged with a subsequent high dose of UVC irradiation, they show accelerated repair of UVC-induced DNA adducts and reduced cell kill. The data indicate a protective role of c-Fos induction by triggering an adaptive response pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genome is permanently harmed by endogenous and exogenous insults that lead to the generation of DNA damage. Fortunately, various DNA repair mechanisms have evolved [1, 2], which are activated by genotoxic stress repairing almost every type of DNA lesion [3]. For example, ultraviolet (UVC) light increases the expression of the DNA repair proteins DDB2, XPC, Pol I, Lig1, and Fen1 [4–8], which are involved in nucleotide excision repair (NER).
An important protein implicated in the protection against genotoxic stress is c-Fos. The c-Fos protein forms together with a member of the Jun family or ATF1 the heterodimeric activator protein 1 (AP-1), a key regulator of gene activity [9, 10]. AP-1 stimulates a broad spectrum of genes harboring consensus sequences in the promoter. c-fos mRNA is immediate-early inducible upon transcriptional activation by growth factors [11], heavy metals [12], UVC light [13], alkylating agents [14], and other genotoxins [15]. Surprisingly, although c-Fos has been extensively analyzed, having a great impact on tumor development (for review see [16]), it “still retains a lot of its mystery” [17]. Thus, whereas regulation of c-Fos expression by growth factors is well established, the regulation by genotoxic stress is still unclear. Besides its impact on tumor development, c-Fos also impacts sensitivity to genotoxic stress. Mouse fibroblasts deficient in c-Fos are more sensitive to UVC light and other chemical DNA damaging agents than the corresponding wild-type [18–21]. We showed that this effect of c-Fos on cellular sensitivity is mediated via regulation of DNA repair. Thus, c-Fos-deficient (p53-proficient) primary rodent fibroblasts display a defect in the repair of cyclobutane pyrimidine dimers (CPDs) because of impaired re-synthesis of XPF [22].
XPF is one of the two endonucleases involved in NER. Whereas the 3′ incision is performed by XPG [23, 24], the 5′ incision is executed by the XPF-ERCC1 complex [25]. The 3′ incision has been reported to be independent of XPF/ERCC1 and is a prerequisite for the 5′ incision [26], which explains why XPF defective cells are able to generate 3′ but not 5′ incision next to the lesion [26]. Upon UVC exposure the expression of xpf mRNA and XPF protein in mouse fibroblasts is strongly reduced. Whereas wild-type cells recover quickly from the transcription block and re-synthesize xpf mRNA, c-fos−/− cells are unable to do so [22]. In c-fos−/− cells this finally leads to decreased repair of UVC-induced CPDs, persistence of NER-intermediate DNA single-strand breaks, prolonged UVC-induced block of replication and transcription, and enhanced Fas-mediated apoptosis [27]. This shows that in mouse fibroblasts c-Fos is involved in the reconstitution of the original xpf gene activity that was repressed upon genotoxic treatment, suggesting a novel concept for the biological function of the “classical” cellular immediate-early genotoxic response, namely stimulation of re-expression of DNA repair genes upon DNA damage and fast restoration of normal DNA repair capacity.
Since responses observed in mouse cells cannot be necessarily translated to human cells, the question arose whether also in human cells c-Fos is involved in the genotoxic stress response via induction of NER genes. Here, we demonstrate for the first time that in human fibroblasts c-Fos plays a decisive role in the induction of XPF and, in part, also of XPG. Induction of c-Fos protein following UVC is postponed and sustained. This causes late stimulation of XPF and XPG synthesis, which enhances the repair of CPDs. We also show that overexpression of c-Fos in human cells, which mimics the situation in tumors, accelerates the re-synthesis of XPF and enhances the repair of UVC-induced DNA damage. In addition, we demonstrate that pre-exposure of cells to a low UVC dose accelerates the repair of CPDs induced by a subsequent challenge dose, which is reminiscent of an adaptive response to DNA damage.
Materials and methods
Cell lines
The human diploid VH10tert foreskin fibroblast cell line immortalized by stable transfection with the telomerase gene (TERT) was kindly provided by Prof. Mullenders (Department of Toxicogenetics at Leiden University Medical Centre, The Netherlands). The human GM637 fibroblast cell line was immortalized by transfection with the SV40 large T-antigen. The mycoplasma-free cells were grown in Dulbecco’s minimal essential medium (DMEM) containing 10% fetal bovine serum (FBS) in 7% CO2 at 37°C.
UVC exposure and treatment with kinase inhibitors
Growth medium was removed and cells were irradiated in the absence of medium with UVC light (wavelength 254 nm) at a dose rate of 1 W/m2 with a radium NSE 11-270 low pressure UVC lamp (Philips, Hamburg, Germany) without fluorescent coating on the bulb surface. The fluence rate was measured by the UV-Radiometer 206 PRC (Krochmann GmbH, Berlin, Germany). Thereafter, the conditioned medium was returned to the dishes, and cells were incubated at 37°C for the appropriate time periods. The JNK1/2/3 inhibitor SP600125 and the p38K inhibitor SB203580 were purchased from Sigma-Aldrich (Hamburg, Germany); the MEK1/2 inhibitor UO126 was from Promega (Mannheim, Germany). The inhibitors were added to the medium at a concentration of 10 μM either 1 h prior to UVC exposure or 16 h after UVC treatment, and remained in contact with the cells until the harvest 24 h after UVC exposure.
Preparation of protein extracts and Western blot analysis
Whole-cell and nuclear extracts were prepared as described previously [28]. Mouse anti-c-Fos (2G9C3) mAb (Abcam, Cambridge, MA, USA), rabbit anti-c-Fos (4) pAb (Santa Cruz Biotechnology, Heidelberg, Germany), anti-FosB (102) pAb (Santa Cruz Biotechnology), anti-Fra-1 (N-17) pAb (Santa Cruz Biotechnology), mouse anti-XPF (Ab-5, clone 51) mAb (NeoMarkers, Thermo Fisher Scientific, Dreieich, Germnany), anti-XPG (clone 8H7) mAb (Santa Cruz Biotechnology), anti-ERCC1 (Ab-1, clone 3H11) mAb (Neomarkers) and β-actin (C4) mAb (Santa Cruz Biotechnology) were diluted 1:500–1:1,000 in 5% non-fat dry milk, 0.1% Tween-TBS and incubated overnight at 4°C. Rabbit anti-ERK2 pAb (Santa Cruz Biotechnology) was diluted 1:3,000 and incubated for 2 h at RT. The protein-antibody complexes were visualized by ECL (Amersham, GE Healthcare, Munich, Germany). For Western blot analysis with phospho-specific antibodies, cells were directly lyzed in 1× SDS-PAGE sample buffer and subsequently sonified. Rabbit phospho-specific as well as non-phosphorylated anti-JNK, anti-p38K and anti-ERK1/2 pAb (Cell Signaling Technology, Boston, MA) were diluted 1:1,000 in 5% BSA/Tween-TBS and rinsed with 0.1% Tween-TBS. The protein-antibody complexes were detected by ECL (Amersham).
Preparation of nuclear extracts and EMSA
Nuclear extracts were prepared as described previously [29] and subjected to electromobility shift assay (EMSA). The sequence of the oligonucleotides specific for the AP-1 binding site of the mouse collagenase promoter was 5′-AGTGGTGACTCATCACT-3′ and the oligonucleotide sequences specific for the AP-1 binding site of the human xpf promoter were xpf-5′-GTAAAAGAATATGAAACATCATGTTCAG, or in case of the mutated AP-1 binding site xpf-mut-5′-GTAAAAGAATATTAAAAATCATGTTCAG. For supershift experiments, 3 μl of antibodies specific for c-Jun (sc45, Santa Cruz), c-Fos (pAb, Active Motif) or p53 (sc100, Santa Cruz) were pre-incubated with 8 μg protein extract for 20 min at room temperature.
Preparation of RNA, RT-PCR and real-time RT-PCR
Total RNA was isolated using the RNA II Isolation Kit (Machery and Nagel, Düren, Germany). One microgram RNA was transcribed into cDNA (Verso cDNA Kit, Thermo Scientific, Bonn, Germany) in a volume of 40 μl, and 3 μl was subjected to RT-PCR performed by the use of specific primers (MWG Biotechnology; Supplement Table 1) and Red-Taq Ready Mix (Sigma-Aldrich). Real-time PCR was performed using the LightCycler FastStart DNA Master SYBR Green I Kit (Roche Diagnostics) and the light cycler of Roche Diagnostics.
Downmodulation of c-Fos
c-Fos-specific siRNA (c-Fos siRNA (h): sc-29221, Santa Cruz Biotechnology) represents a pool of four target-specific 20–25 nt siRNAs. VH10tert cells were transfected as described [30].
Southwestern blot analysis
Genomic DNA was isolated by the use of the QIA(amp) blood mini kit (Qiagen, Hilden, Germany), and Southwestern blot analysis was performed as reported [31]. Monoclonal antibodies against UVC-induced DNA damage, anti-CPDs (clone TDM-2) and (6-4)PPs (clone 64 M-2) were purchased from Cosmo Bio Co., Ltd. (Tokyo, Japan) and were diluted 1:200 in 5% milk/0.1% Tween-TBS and incubated overnight at 4°C. The additional Western blot procedure and detection were performed as described above.
Quantification of Western and Southwestern blots
The signals were quantified using the GeneTool software and InGenius Gel Documentation System from Synngene (Cambridge, UK).
Cloning of the c-fos cDNA and generation of stable clones
The c-fos cDNA was cloned by RT-PCR amplification using specific primers (c-fos-cDNA-up, GATGATGTTCTCGGGCTTCAACGC, c-fos-cDNA-low: CTCACAGGGCCAGCAGCGTGGGTG) and cloned into the pcDNA3.1/V5-His-Topo (Invitrogen, Darmstadt, Germany). The recombinant vectors were transfected into GM637 cells using Effectene reagent (Qiagen).
Determination of apoptosis
To monitor UVC-induced apoptosis, cells were incubated for 30 min with 0.1 mg/ml RNase in PBS and stained with propidium iodide (PI), and the sub-G1 fraction was determined by flow cytometry as described previously [32]. Experiments were repeated at least three times, mean values ± SD are shown, and data were compared statistically using Student’s t test.
Results
Induction of c-Fos by UVC light in human fibroblasts
To analyze the UVC-mediated regulation of c-Fos and XPF in human cells, we utilized the human fibroblast cell line VH10tert. It is important to note that VH10tert cells are diploid (data not shown), although they are immortalized by TERT. Most, if not all, immortalized cells are aneuploid. The same is true for human tumor cells and cultured mouse fibroblasts, which tend to acquire genetic alterations and in most cases are highly aneuploid. This might change the phenotype of the cells and can alter the DNA damage response. For this reason we decided to work with the cell line VH10tert. VH10tert cells are p53 wild type (Supplement 1A) and show induction of ddb2 and xpc upon UVC exposure (Supplement 1B). Since p53-deficient mouse and human fibroblasts are defective in NER [33, 34], the p53 status was routinely checked. VH10tert cells are proficient in the repair of CPDs and (6-4)PPs (Supplement 1C) and do not die after exposure to doses below 10 J/m2 UVC (Supplement 1D). Similar to mouse fibroblasts [35], the induction of c-fos mRNA is biphasic. A strong but transient induction was observed 1 h after UVC exposure (10 J/m2) of VH10tert cells. The RNA returned to basal level 1 h later (Fig. 1a, left panel). A second wave of induction was observed 4 h after UVC exposure and reached maximal levels 16–24 h thereafter (Fig. 1a, right panel). Interestingly, the same was observed for fosB, but not for the third member of the Fos family, fra1, which was not induced at all. In contrast to c-fos and fosB mRNA induction, no immediate-early upregulation of the corresponding proteins was observed in VH10tert cells, whereas a late and long-lasting upregulation of c-Fos and FosB protein occurred 18–48 h after exposure (Fig. 1b). In contrast to c-Fos and FosB, which showed no basal expression, Fra1 was expressed at a high level in untreated cells, and its expression remained unaffected following UVC exposure (Fig. 1b).
UVC-induced activation of the MAPK cascade and induction of Fos proteins. a Exponentially growing VH10tert cells were exposed to 10 J/m2 UVC. At different time points after exposure, total RNA was isolated and semi-quantitative RT-PCR was performed using c-fos, fosB, fra1 or, as loading control, gapdh specific primers. b, c Exponentially growing VH10tert cells were exposed to 10 J/m2 UVC for the indicated times. Protein extracts were prepared and subjected to Western blot analysis. The filter was incubated with c-Fos, FosB or Fra1 specific antibodies (b) or incubated with p-JNK, JNK, p-p38K, p38K, p-ERK1/2 and ERK1/2 specific antibodies (c); the two phosphorylated JNK specific bands (JNK1 p46 and JNK2 p54) are labeled by arrows. d Left panel: Exponentially growing VH10tert cells were exposed to 10 J/m2 UVC; 16 h later 10 μM of a specific inhibitor for JNK1/2/3 (SP600125), p38K (SB103580), and MEK1/2 (UO126), respectively, was added. Additional 8 h later, protein extracts were prepared and subjected to Western blot analysis. The filter was incubated with c-Fos and ERK2 specific antibodies. d Right panel: Exponentially growing VH10tert cells were pre-incubated for 1 h with a specific inhibitor for JNK, p38K and MEK1/2, respectively. Thereafter cells were exposed to 10 J/m2 UVC, and the conditioned medium containing the inhibitor was re-added. Then 24 h later protein extracts were prepared and subjected to Western blot analysis. The filter was incubated with c-Fos and ERK2 specific antibodies
Activation of c-Fos by stress-inducible protein kinases
Previous studies showed that stress-inducible kinases such as c-Jun N-terminal kinase (JNK) and mitogen-activated protein kinases [MAPKs; extracellular signal-regulated protein kinases (ERKs)] are involved in the induction and activation of c-Fos in human cells depending on the cell type and the stimulus. To analyze which of the protein kinases are involved in UVC-mediated induction of c-Fos in human fibroblasts, we analyzed the activation of JNK, p38 kinase (p38K) and ERK1/2 using phospho-specific antibodies. JNK and p38K were not active in untreated cells. They became phosphorylated 1 h after UVC treatment and remained phosphorylated for at least 8 h (Fig. 1c). Thereafter, the amount of phosphorylated kinases declined, but was still detectable up to 32 h after exposure. In contrast, ERK1/2 was already activated in untreated cells, and phosphorylation was not enhanced by UVC exposure (Fig. 1c). Since the expression of c-Fos was detected 18 h post-UVC, the question arose whether the low remaining amounts of active JNK or p38K might still be associated with c-Fos activation. To analyze this we utilized different kinase-specific inhibitors, which were added to the medium either 16 h after or 1 h prior to UVC irradiation. Cells were harvested 24 h after UVC. As shown in Fig. 1d, inhibition of the early JNK activity by the JNK1/2/3 inhibitor SP600125 (left panel) or the early and late ERK1/2 activity using UO126 (an inhibitor of the upstream MEK1/2; both panels) resulted in significant abrogation of c-Fos expression, whereas inhibition of p38K by SB203580 had no effect on c-Fos expression.
Induction of the NER endonucleases XPF and XPG upon UVC exposure
Next, we analyzed whether the expression of the NER proteins ERCC1, XPF, and XPG is elevated upon UVC. Similar to data obtained in mouse fibroblasts, in human cells xpf and xpg mRNAs were highly unstable and downregulated after UVC exposure [shown by semi-quantitative RT-PCR (Fig. 2a) and real-time RT-PCR (Fig. 2b)]. Thus, upon treatment with 10 J/m2 the xpg mRNA level dropped below 50% control level, and the xpf expression was nearly undetectable 4–8 h upon exposure. Thereafter, both mRNAs were re-synthesized, exceeding the control level 24–48 h after exposure. Similar results were observed by microarray analysis (using a self-designed DNA repair array) showing induction of the NER genes xpf, xpg, ddb2 and xpc 32 h after exposure to 10 J/m2 UVC (data not shown). The enhanced mRNA expression resulted in elevated expression levels of XPF and XPG (Fig. 2c). Starting 24 h after exposure to 10 J/m2 UVC, increased levels of XPF and XPG proteins were observed, exceeding the basal expression for at least 24 additional hours, clearly indicating that both proteins are induced upon UVC exposure. The expression of ercc1 mRNA and the corresponding ERCC1 protein was only marginally altered following UVC.
UVC-mediated induction of NER proteins. a Exponentially growing VH10tert cells were exposed to 10 J/m2 UVC. At different time points after exposure, total RNA was isolated and semi-quantitative RT-PCR was performed using xpf, xpg, ercc1 or, as loading control, gapdh specific primers. b Exponentially growing VH10tert cells were exposed to 10 J/m2 UVC for the indicated times. Total RNA was isolated and real-time RT-PCR was performed using xpf, xpg, ercc1 or, as loading control, gapdh specific primers. For quantification, the expression was normalized to gapdh and the untreated control was set to 1. Data are the mean of three independent experiments ± SD. c Exponentially growing VH10tert cells were exposed to 10 J/m2 UVC for the indicated time points. Protein extracts were prepared and subjected to Western blot analysis. The filter was incubated with XPF, XPG or ERCC1 specific antibodies. β-actin, loading control
Knockdown of c-Fos abrogates XPF induction and CPD removal following UVC and enhances apoptosis
To further substantiate the role of c-Fos in the regulation of re-synthesis of XPF, siRNA experiments were performed. As shown in Fig. 3a, transfection of VH10tert cells with c-Fos specific siRNA reduced the UVC-induced expression of c-Fos by more than 90%. Downmodulation was observed 24–48 h upon siRNA transfection. Knockdown of c-Fos had a pronounced effect on the expression of the NER endonuclease XPF. Thus, in non-transfected and nonsense-siRNA (ns-si) transfected cells, UVC light strongly induced XPF expression, as determined 32 h post-UVC, whereas this induction was completely abrogated after transfection with c-Fos specific siRNA (si) (Fig. 3b). Similar to XPF, the induction of XPG was also reduced under c-Fos knockdown conditions (Fig. 3b). This effect was, however, not as pronounced as for XPF, indicating that besides c-Fos other factors play a role in the regulation of UVC-mediated XPG induction. In addition, knockdown of c-Fos significantly reduced the removal of CPDs (Fig. 3c) and sensitized cells to the apoptosis-inducing effect of UVC light (Fig. 4), indicating that re-synthesis and induction of XPF are crucial events in the repair of UVC-induced DNA damage in human fibroblasts. Removal of (6-4)PPs was not affected by c-Fos knockdown (Fig. 3d). The repair of (6-4)PPs occurred early after irradiation (<8 h), at a time when neither c-Fos nor XPF are induced. These data go along with the fact that in contrast to CPDs, (6-4)PPs are efficiently recognized and processed by NER (for review see [36]), and XPF appears not to be a limiting factor.
Effect of c-Fos knockdown on XPF induction and NER capacity. a Exponentially growing VH10tert cells were transfected with c-Fos specific siRNA (si) or nonsense-siRNA (ns-si) and 24 h later exposed to 10 J/m2 UVC. Protein extracts were prepared 24 or 48 h after exposure and subjected to Western blot analysis. The filter was incubated with c-Fos specific antibodies. β-actin was used as loading control. b Exponentially growing VH10tert cells were transfected with c-Fos specific siRNA (si) or nonsense-siRNA (ns-si) and 24 h later exposed to 10 J/m2 UVC. Protein extracts were prepared 32 h after exposure and subjected to Western blot analysis. The filter was incubated with XPF or XPG specific antibodies. β-actin was used as loading control. The induction factor (IF) is derived from densitometric measurement of XPF and XPG signal and normalized to β-actin expression. c, d To analyze NER capacity upon c-Fos knockdown VH10tert cells were transfected with c-Fos specific siRNA (si) or nonsense-siRNA (ns-si) and 24 h later exposed to 10 J/m2 UVC. At different time points following irradiation, genomic DNA was isolated; equal amounts of DNA were blotted and subjected to incubation with c anti-CPD antibodies or d anti-(6-4)PP antibodies (con, non-exposed control). Representative blots are shown (left panel) and the mean data of three independent experiments ± SD (right panel). The CPD signal determined 5 min after treatment was set to 100%. Data were compared statistically using Student’s t test (**p < 0.01)
Effect of c-Fos knockdown on sensitivity to UVC light. To monitor the impact of c-Fos knockdown on sensitivity to UVC light, VH10tert cells were transfected with c-Fos specific siRNA (c-fos-si) or nonsense-siRNA (non-si); 48 h later, cells were irradiated with 10 (a) or 25 J/m2 UVC (b) and incubated for 96 or 144 h. Cells were stained with PI, and the sub-G1 fraction (apoptotic cells) was determined by flow cytometry. Experiments were repeated at least three times, mean values ± SD are shown, and data were compared statistically using Student’s t test (*p < 0.05, **p < 0.01)
Overexpression of c-Fos accelerates XPF induction and enhances resistance to UVC
To further prove the importance of c-Fos in the regulation of XPF expression and resistance to UVC light, we transfected human GM637 fibroblasts with human c-fos cDNA. Untransfected GM637 cells showed a c-Fos response comparable to VH10tert cells, with sustained induction of the c-fos mRNA 4–16 h post-UVC (Supplement 2A) and of the c-Fos protein 18 h post-UVC (Supplement 2B). In addition, UVC-induced activation of JNK and p38K was also observed (Supplement 2C). Stably transfected c-Fos clones were selected that showed enhanced expression of c-fos mRNA and c-Fos protein (A4, B4, B5, and D1) along with the parental GM637 cells and a clone (A2) not overexpressing c-Fos (Fig. 5a). It is important to note that all clones showed nearly identical proliferation rates (Supplement 3A), which therefore cannot account for possible differences in DNA repair and/or sensitivity to UVC light. In spite of overexpression of c-Fos, the basal expression of xpf and xpg remained equal in all clones (Fig. 5b). However, a strong difference was observed in the UVC-mediated induction of xpf (Fig. 5c, left panel and Supplement 3B). In GM637 cells and the clone A2, reduction of xpf expression was observed 8–16 h after exposure, followed by re-synthesis and induction after 24–32 h. In contrast, the c-Fos overexpressing clones (B4, B5, and D1) showed no reduction in xpf mRNA and an accelerated induction already 8–16 h post-UVC. Interestingly, the clone A4, which displayed only weak c-Fos overexpression, showed an intermediate phenotype with no reduction in xpf mRNA and upregulation 24–32 h after UVC. The differences between c-Fos non-expressing and overexpressing clones were not that pronounced for xpg. Thus, in no case more than a two-fold induction was observed (Fig. 5c, right panel and Supplement 3B). Overexpression of c-Fos also affected removal of CPDs and resistance to UVC light. Whereas in GM637 cells and the clone A2, used as an empty vector control, CPD removal was observed 48–72 h after UVC, the repair was significantly accelerated in the c-Fos overexpressing clones A4, B4, B5, and D1, removing CPDs 32–48 h thereafter (Fig. 5d). In accordance with this, the enhanced repair of CPDs reduced the sensitivity to UVC light. Thus, 72 h post-UVC GM637 cells and the clone A2 showed ~30% apoptosis, whereas the frequency of apoptosis in all c-Fos overexpressing clones was <15% (Fig. 5e).
Effect of c-Fos overexpression on XPF induction, NER capacity and apoptosis. a Human GM637 fibroblasts were transfected with a c-Fos expression plasmid. Parental GM637 cells and five clones expressing different amounts of c-Fos were selected. Total RNA and protein extracts were prepared from the individual clones and subjected to RT-PCR (upper panel) or Western blot analysis (lower panel). The expression of c-Fos was analyzed via c-Fos specific primers or antibodies. For loading control, gapdh or β-actin was detected. b Total RNA was prepared from the individual clones and subjected to RT-PCR. The expression of xpf and xpg was analyzed via specific primers. For loading control, gapdh was detected. c Exponentially growing clones were exposed to 10 J/m2 UVC. Total RNA was isolated at different time points after exposure and real-time RT-PCR was performed using xpf (left panel) or xpg (right panel) specific primers. For quantification, the expression was normalized to gapdh and the untreated control was set to 1. d To analyze NER capacity upon c-Fos overexpression, the different clones were exposed to 10 J/m2 UVC. At different time points following irradiation, genomic DNA was isolated, and equal amounts of DNA were blotted and incubated with anti-CPD antibodies (con, non-exposed control). e To monitor the impact of c-Fos overexpression on sensitivity to UVC light, the different cell clones were irradiated with 10 J/m2, and 96 h later cells were stained with PI and the sub-G1 fraction (apoptotic cells) was determined by flow cytometry. Experiments were repeated at least three times, mean values ± SD are shown, and data were compared statistically using Student’s t test (*p < 0.05, **p < 0.01)
Binding of AP-1 to the human xpf promoter
Computer-based analysis of the human xpf promoter (using the program PATCH 1.0; http://www.gene-regulation.com) revealed the presence of a putative AP-1 binding sequence (ATGAAAC). Electromobility shift assays (EMSA) were performed using radioactively labeled oligonucleotides harboring either the putative AP-1 binding sites of the xpf promoter or, for control, the AP-1 binding site of the collagenase promoter (mmp1). The oligonucleotides were incubated with nuclear extracts obtained from VH10tert cells, either untreated or treated with 10 J/m2 UVC. As shown in Fig. 6a, time-dependent induction of AP-1 binding activity was observed using the mmp1 AP-1 binding site or the putative xpf AP-1 binding site. Competition experiments with non-radioactively labeled oligonucleotides containing the AP-1 sequence of the mmp1 promoter, but not with non-radioactively labeled oligonucleotides containing the p53 binding site of the p21 promoter, abrogated the recognition of the xpf promoter by AP-1 (Fig. 6b). Mutation of two nucleotides in the xpf AP-1 binding site (xpf-mut, ATTAAAA) completely abrogated the recognition by AP-1, showing the specificity of the binding complex (Fig. 6b). To further substantiate the specificity of the AP-1 binding and to identify binding partners for c-Fos involved in the recognition of the xpf specific AP-1 site, supershift experiments were performed. Both the collagenase AP-1 site (mmp1 AP-1) and the xpf specific AP-1 site (xpf AP-1) were recognized by c-Fos and c-Jun, but not by p53, which was included as negative control (Fig. 6c).
Identification of AP-1 binding sites within the xpf promoter. a Oligonucleotides containing either the AP-1 binding site of the collagenase promoter (mmp1) or the AP-1 binding sites of the xpf promoter were incubated with nuclear extracts from VH10tert cells exposed to 10 J/m2 UVC light and harvested at different time points later and subjected to EMSA. The specific complex is indicated by an arrow. b For verifying the specificity of the reaction, a 5-, 10-, or 25-fold excess of unlabeled oligonucleotides containing either the p53 binding site of p21 (p21) or the AP-1 binding site of the collagenase promoter (mmp1) was added to the assay using labeled xpf promoter specific oligonucleotides. In addition EMSA was performed using point-mutated oligonucleotides (containing two exchanges in the xpf specific AP-1 binding site; xpf-mut) and nuclear extracts from VH10tert cells exposed to 10 J/m2 UVC light and harvested 8 and 24 h later. c EMSA supershift assay. Composition of the AP-1 factor bound to the mmp1 AP-1 and xpf AP-1 oligonucleotide was analyzed by the addition of specific antibodies against p53, c-Jun, or c-Fos to the reaction. The supershifted complex is indicated by an arrow
UVC-triggered induction of XPF provokes an adaptive response
Under “normal” biological conditions, cells are confronted with low but constant genotoxic insults, and it is conceivable that cells respond with the induction of protective functions. Therefore, the question arose whether a pretreatment dose, which is effective in increasing DNA repair activity, can reduce the genotoxic effect of a subsequent challenge dose. We checked this by pretreating VH10tert cells with a dose of 5 J/m2 UVC to induce c-Fos, which was shown to upregulate XPF/XPG mediated NER activity. Thirty-two hours later non-preexposed and preexposed cells were challenged with a dose of 10 J/m2 UVC, and several time points later the remaining CPDs were determined. As shown in Fig. 7a, 32 h after pretreatment (con, preexposed) CPDs were still detectable, indicating that they did not get completely repaired. Obviously, the subsequent exposure to the challenge dose resulted in a higher level of CPDs compared to non-preexposed cells (0.1 h after subsequent exposure). However, despite the higher level of CPDs in the preexposed cells, the remaining CPDs after an additional 16, 24, and 32 h were comparable between preexposed and non-preexposed cells (Fig. 7a). This indicates that CPD repair in the preexposed cells was enhanced. In contrast to the repair of CPDs, no effect was observed as to the removal of (6-4)PPs (Fig. 7b). The quantification of the repair kinetics is shown in the bar diagrams (Fig. 7a, b, right panel).
Adaptive response to UVC-induced damage. a, b VH10tert cells were either not pre-exposed (control) or pre-exposed to 5 J/m2 UVC. Thirty-two hours later the cells were exposed to 10 J/m2 UVC. At different time points later, genomic DNA was isolated, and equal amounts of DNA were blotted and subjected to incubation with anti-CPD (a) or anti-(6-4)PP antibodies (b) (con, non-exposed control). The representative experiment is shown in the left panel. The right panel shows the mean remaining CPDs in three independent experiments ± SD. c To monitor the impact of pre-exposure on sensitivity to UVC light, VH10tert cells were either pre-exposed or not pre-exposed to 5 J/m2 UVC and 32 h later exposed to 25 J/m2 UVC. One hundred twenty hours later, cells were stained with PI, and the sub-G1 fraction (apoptotic cells) was determined by flow cytometry. In addition, cells were irradiated with 5 J/m2 and incubated for 152 h. Experiments were repeated at least three times, mean values ± SD are shown, and data were compared statistically using Student’s t test (*p < 0.05)
Enhanced NER triggered by a low UVC dose might impact on the killing response of the cells following a subsequent challenge dose. To prove this, we preexposed VH10tert cells to 5 J/m2 UVC and challenged them 32 h later with a dose of 25 J/m2. As shown in Fig. 7c, unexposed VH10tert cells (con) showed a basal apoptosis frequency of 11%, and cells exposed to 5 J/m2 UVC showed 16% apoptosis. Cells exposed to 25 J/m2 UVC displayed a frequency of 34% apoptosis, and cells that were preexposed to 5 J/m2 UVC and challenged with 25 J/m2 UVC showed a significantly reduced apoptosis frequency of 26% (Fig. 7c). We should note that the rate of CPD removal could theoretically depend on the overall level of CPDs. In this case, higher overall amounts of lesions might enhance the kinetics of CPD removal. As shown in Supplement 1C, the repair capacity indeed depends on the levels of CPDs. However, higher CPD levels are repaired worse in comparison to low levels. In the case of preexposure with UVC, higher CPD levels are repaired better. This clearly argues against the possibility that an increased overall amount of lesions explains the increased repair in UVC-preexposed cells. Therefore, we conclude that stimulation of NER by a low UVC dose protects against cell death resulting from a subsequent UVC challenge dose.
Discussion
Mouse fibroblasts deficient for c-Fos are hypersensitive to the cytotoxic, apoptosis-inducing, and clastogenic effects of UVC light [18, 19, 21] and other genotoxins [20]. The reason for the hypersensitivity remained enigmatic for more than 10 years, until we demonstrated that c-Fos/AP-1 plays a key role in regulating the expression of XPF upon UVC exposure [22]. Thus, in mouse fibroblasts UVC exposure induces a strong reduction in the expression of xpf mRNA and XPF protein. The re-synthesis of XPF is triggered by c-Fos/AP-1, leading to reconstitution of XPF expression and enhanced DNA repair activity. In the absence of c-Fos, cells are unable to remove CPDs from the DNA and undergo apoptosis at high frequency via the Fas pathway. The apoptotic program becomes activated by prolonged transcription blockage leading to a reduced expression of MKP1, sustained JNK activation, and stimulation of synthesis of the Fas ligand [27].
Here, we analyzed the UV response in human fibroblasts. Notably we wished to elucidate whether also in human cells c-Fos induces XPF expression and affects resistance to UVC light. We utilized UV light of a wavelength of 254 nm (UVC), which induces predominantly CPDs and (6-4)PPs, whereas UVB and UVA produce a mixture of these lesions and reactive oxygene species, which could superimpose the effects of c-Fos mediated NER regulation. To mimic the dose level of the “normal” exposure situation, where only wavelengths above 280 nm reach the surface of the earth, we performed all experiments using 10 J/m2 UVC. A dose of 10 J/m2 UVC induces about 0.3 CPDs per kbp. This is comparable to about 80 kJ/m2 sunlight or 1 kJ/m2 UVB [37]. It was reported that 0.062 CPDs are induced during a 30-min sunbath in August in Paris. This corresponds to a sun exposure period of 2.5 h [38], yielding a CPD level comparable to 10 J/m2 UVC. This calculation indicates that the UVC dose used is relevant for the in vivo situation. Further we should note that UVC represents a paradigm for a genotoxin that induces DNA lesions repaired by NER.
First, we observed in human fibroblasts a biphasic c-fos mRNA induction similar to what was reported for mouse cells [35]. However, in human cells the first wave of mRNA, which was observed ~60 min after UVC exposure, was not accompanied by a detectable increase in the c-Fos protein level. Therefore, the physiological role of this immediate-early c-fos mRNA induction, which is supposed to be triggered by EGFR activation [39], remains unclear. More important appears to be the late and sustained upregulation of the c-fos mRNA level, reaching a maximum 16–24 h after UVC treatment that was accompanied by an increase in the c-Fos protein level. Whether the second wave of c-fos mRNA induction is caused by transcriptional upregulation or mRNA stabilization [35] will be the subject of forthcoming studies.
It has been shown that upon UVC treatment JNK1/2 phosphorylates ELK1, which transcriptionally activates c-Fos [40], whereas p38K phoshorylates c-Fos at Thr325, leading to nuclear translocation and transcriptional activation of c-Fos [41]. In human fibroblasts we showed that inhibition of early JNK activity and inhibition of the early and late ERK1/2 expression results in decreased c-Fos expression following UVC. Inhibition of p38K had no effect at all. The data suggest that the early activation of JNK is the initial trigger for the UVC-induced c-Fos synthesis. However, once c-Fos protein is expressed not only JNK, but also ERK1/2 appears to be responsible for its sustained upregulation.
Similar to mouse fibroblasts [22], in human fibroblasts UVC exposure leads to a reduced expression level of xpf and xpg mRNA. However, whereas in the mouse system c-Fos mediates the restoration of the “normal” XPF expression, in human fibroblasts c-Fos activation leads to a strong induction (i.e., overexpression) of the xpf and xpg mRNA and the corresponding proteins. The induction of XPF in human cells is completely dependent on the presence of c-Fos since knockdown of c-Fos abrogates XPF induction, reduces removal of CPDs from the DNA, and sensitizes cells to UVC as revealed by an increased frequency of apoptosis. The effect of c-Fos knockdown on XPG induction was not as pronounced as for XPF, indicating that, apart from c-Fos, other transcription factors are involved in the upregulation of XPG.
The finding that xpf is a target of AP-1 was confirmed by the detection of an AP-1 binding site in the human xpf promoter, which was recognized in vitro by AP-1. To further prove the role of c-Fos in the regulation of XPF and the repair of UVC-induced DNA damage, c-Fos was stably overexpressed in human GM637 cells. All c-Fos overexpressing clones showed the same basal expression of xpf and xpg. However, marked differences were observed upon UVC exposure. In c-Fos overexpressing cells treated with UVC the initial reduction of xpf mRNA was not observed. The late induction was accelerated, showing its maximum already 16 h after UVC treatment. Interestingly clone A4, which shows a lower c-Fos expression than the other c-Fos overexpressing clones (B4, B5, B1), showed an intermediate response concerning xpf induction (Fig. 5c), but the same repair and survival rate as the other clones, indicating that counteracting the xpf transcriptional inhibition below the control level is sufficient for eliciting UVC protection. This is similar to our previous data obtained with mouse cells where c-Fos triggered the re-synthesis of XPF without inducing it above the control level.
To ascertain whether induction of XPF leads to accelerated DNA repair, we analyzed the repair of DNA lesions in c-Fos overexpressing cells. Indeed, these cells, which were clonal derivatives of stable c-Fos transfectants, showed an accelerated removal of CPDs and a reduced apoptotic response following UVC. It is important to note that overexpression of c-Fos had no effect on cell proliferation, which was shown to influence the cell’s sensitivity to UVC [42]. An explanatory model compiling also data from the literature is shown in Fig. 8. Constitutive c-Fos overexpression might mimic the situation in cancer, as several tumors overexpress c-Fos. This might be a hurdle for clinical settings using anticancer drugs that induce bulky lesions such as cisplatin [43].
Model of MAPK, c-Fos, and XPF expression under physiological and c-Fos overexpressing conditions upon UVC exposure. In normal unexposed fibroblasts under physiological conditions (upper flow chart) neither c-fos mRNA nor c-Fos protein is expressed. Upon UVC exposure MAPKs are activated and phosphorylate various transcription factors such as Elk1 and c-Jun, which are responsible for the transcriptional activation of c-fos mRNA. This is followed by expression of c-Fos protein, which in turn becomes activated via phosphorylation by MAPKs and binds to its partner, e.g., c-Jun, to form the active AP-1 complex, which finally activates the xpf promoter. In UVC-irradiated c-Fos overexpressing cells (lower flow chart) the first steps, namely induction of xpf mRNA and XPF protein, have already occurred because the MAPKs directly activate c-Fos leading to accelerated induction of XPF and faster repair of CPDs. Although overexpressed in unexposed cells, c-Fos is not transcriptionally active because it needs activation by MAPKs and a binding partner such as c-Jun. Following UVC exposure these proteins are activated, c-Fos containing AP-1 is formed, and target genes including xpf are induced
c-Fos is an important factor in tumorigenesis [44], although due to its multiple functions [16, 17] the molecular basis for the tumorigenic effect of c-Fos is not yet fully understood. Theoretically, induction of c-Fos or permanent overexpression and sustained activation could lead to an increase in repair activity and thereby an increase in the resistance of transformed cells to genotoxic insults. This might allow the cells to survive even when they were genetically damaged, which could cause selection advantage of transformed cells, thus stimulating tumorigenesis. Not only overexpression of c-Fos but also decreased expression was reported in tumor cells. An example is basal cell carcinomas where c-fos mRNA expression was markedly reduced [45]. In this case, decreased XPF expression and NER activity might lead to an accumulation of bulky DNA lesions that are processed by translesion synthesis. This would lead to enhanced mutation rates and thereby accelerated tumorigenicity. Although rather speculative, the hypothesis is conceivable and awaits experimental verification.
Since the induction of XPF is a rather late event upon UVC exposure, a question concerning the biological relevance arises. As we showed, induction of XPF not only affects the response to the initial UVC damage, but also impacts CPD removal and survival of cells after a subsequent UVC challenge dose. It seems that at this time point the cells have been conditioned via the induction of XPF, thus increasing the DNA repair capacity of CPDs. This, however, is not the case for (6-4)PPs, which are quickly removed under both conditions. Obviously the induction of XPF and XPG is important solely for the removal of lesions that are difficult to detect and repair. It is reasonable to assume that the response is important for protecting cells exposed chronically to UVC light and other genotoxic insults. An intriguing question in this context is whether simulated sunlight, similar to UVC, also provokes an adaptive response in human cells. This will be analyzed in future experiments.
Overall, we show that in human fibroblasts c-Fos induction is a late response to UVC irradiation, which triggers upregulation of XPF and, to a lesser extent, XPG, thereby enhancing the NER capacity and protecting against UVC-induced cell death. Due to an increased repair capacity these “primed” cells gained protection against a second dose of UVC. The data provide evidence for an adaptive response of human cells to UVC light. Future work will show whether this concept can be extended to other DNA damaging agents, including UVB and anticancer drugs, which induce DNA lesions repaired by NER.
Abbreviations
- AP-1:
-
Activator protein 1
- CPDs:
-
Cyclobutane pyrimidine dimers
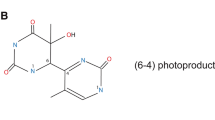
- (6-4)PP:
-
6-4 Photoproducts
- EMSA:
-
Electromobility shift assay
- ERK:
-
Extracellular signal-regulated protein kinase
- JNK:
-
c-Jun N-terminal kinase
- MAPK:
-
Mitogen-activated protein kinase
- MEFs:
-
Mouse embryonic fibroblasts
- mAb:
-
Monoclonal antibody
- NER:
-
Nucleotide excision repair
- pAb:
-
Polyclonal antibody
- siRNA:
-
Small interfering RNA
- UV:
-
Ultraviolet
- wt:
-
Wild-type
References
Christmann M, Tomicic MT, Roos WP, Kaina B (2003) Mechanisms of human DNA repair: an update. Toxicology 193:3–34
Hoeijmakers JH (2009) DNA damage, aging, and cancer. N Engl J Med 361:1475–1485
Christmann M, Fritz G, Kaina B (2007) Induction of DNA repair genes in mammalian cells in response to genotoxic stress. In: D. Lankenau (Ed) Genome dynamics and stability, vol 1. Springer, Berlin, pp 383–398
Adimoolam S, Ford JM (2002) p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc Natl Acad Sci USA 99:12985–12990
Montecucco A, Savini E, Biamonti G, Stefanini M, Focher F, Ciarrocchi G (1995) Late induction of human DNA ligase I after UV-C irradiation. Nucleic Acids Res 23:962–966
Hwang BJ, Ford JM, Hanawalt PC, Chu G (1999) Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci USA 96:424–428
Yang J, Chen Z, Liu Y, Hickey RJ, Malkas LH (2004) Altered DNA polymerase iota expression in breast cancer cells leads to a reduction in DNA replication fidelity and a higher rate of mutagenesis. Cancer Res 64:5597–5607
Christmann M, Tomicic MT, Origer J, Kaina B (2005) Fen1 is induced p53 dependently and involved in the recovery from UV-light-induced replication inhibition. Oncogene 24:8304–8313
Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M (1988) The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell 54:541–552
Rauscher FJ 3rd, Sambucetti LC, Curran T, Distel RJ, Spiegelman BM (1988) Common DNA binding site for Fos protein complexes and transcription factor AP-1. Cell 52:471–480
Muller R, Bravo R, Burckhardt J, Curran T (1984) Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature 312:716–720
Gubits RM, Fairhurst JL (1988) c-fos mRNA levels are increased by the cellular stressors, heat shock and sodium arsenite. Oncogene 3:163–168
Buscher M, Rahmsdorf HJ, Litfin M, Karin M, Herrlich P (1988) Activation of the c-fos gene by UV and phorbol ester: different signal transduction pathways converge to the same enhancer element. Oncogene 3:301–311
Dosch J, Kaina B (1996) Induction of c-fos, c-jun, junB and junD mRNA and AP-1 by alkylating mutagens in cells deficient and proficient for the DNA repair protein O6-methylguanine-DNA methyltransferase (MGMT) and its relationship to cell death, mutation induction and chromosomal instability. Oncogene 13:1927–1935
Hollander MC, Fornace AJ Jr (1989) Induction of fos RNA by DNA-damaging agents. Cancer Res 49:1687–1692
Milde-Langosch K (2005) The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer 41:2449–2461
Durchdewald M, Angel P, Hess J (2009) The transcription factor Fos: a Janus-type regulator in health and disease. Histol Histopathol 24:1451–1461
Lackinger D, Eichhorn U, Kaina B (2001) Effect of ultraviolet light, methyl methanesulfonate and ionizing radiation on the genotoxic response and apoptosis of mouse fibroblasts lacking c-Fos, p53 or both. Mutagenesis 16:233–241
Haas S, Kaina B (1995) c-Fos is involved in the cellular defence against the genotoxic effect of UV radiation. Carcinogenesis 16:985–991
Kaina B, Haas S, Kappes H (1997) A general role for c-Fos in cellular protection against DNA-damaging carcinogens and cytostatic drugs. Cancer Res 57:2721–2731
Lackinger D, Kaina B (2000) Primary mouse fibroblasts deficient for c-Fos, p53 or for both proteins are hypersensitive to UV light and alkylating agent-induced chromosomal breakage and apoptosis. Mutat Res 457:113–123
Christmann M, Tomicic MT, Origer J, Aasland D, Kaina B (2006) c-Fos is required for excision repair of UV-light induced DNA lesions by triggering the re-synthesis of XPF. Nucleic Acids Res 34:6530–6539
O’Donovan A, Davies AA, Moggs JG, West SC, Wood RD (1994) XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371:432–435
Habraken Y, Sung P, Prakash L, Prakash S (1994) A conserved 5′ to 3′ exonuclease activity in the yeast and human nucleotide excision repair proteins RAD2 and XPG. J Biol Chem 269:31342–31345
Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, Rademakers S, de Rooij J, Jaspers NG, Hoeijmakers JH, Wood RD (1996) Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86:811–822
Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J 16:6559–6573
Christmann M, Tomicic MT, Aasland D, Kaina B (2007) A role for UV-light-induced c-Fos: stimulation of nucleotide excision repair and protection against sustained JNK activation and apoptosis. Carcinogenesis 28:183–190
Christmann M, Kaina B (2000) Nuclear translocation of mismatch repair proteins MSH2 and MSH6 as a response of cells to alkylating agents. J Biol Chem 275:36256–36262
Christmann M, Tomicic MT, Kaina B (2002) Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutS{alpha} mismatch-binding activity. Nucleic Acids Res 30:1959–1966
Tomicic MT, Christmann M, Kaina B (2010) Topotecan triggers apoptosis in p53-deficient cells by forcing degradation of XIAP and survivin thereby activating caspase-3-mediated bid cleavage. J Pharmacol Exp Ther 332:316–325
Koberle B, Roginskaya V, Wood RD (2006) XPA protein as a limiting factor for nucleotide excision repair and UV sensitivity in human cells. DNA Repair (Amst) 5:641–648
Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C (1991) A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods 139:271–279
Smith ML, Ford JM, Hollander MC, Bortnick RA, Amundson SA, Seo YR, Deng CX, Hanawalt PC, Fornace AJ Jr (2000) p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol 20:3705–3714
Ford JM, Hanawalt PC (1997) Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem 272:28073–28080
Blattner C, Kannouche P, Litfin M, Bender K, Rahmsdorf HJ, Angulo JF, Herrlich P (2000) UV-induced stabilization of c-fos and other short-lived mRNAs. Mol Cell Biol 20:3616–3625
Costa RM, Chigancas V, Galhardo Rda S, Carvalho H, Menck CF (2003) The eukaryotic nucleotide excision repair pathway. Biochimie 85:1083–1099
Perdiz D, Grof P, Mezzina M, Nikaido O, Moustacchi E, Sage E (2000) Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells. Possible role of Dewar photoproducts in solar mutagenesis. J Biol Chem 275:26732–26742
Kuluncsics Z, Perdiz D, Brulay E, Muel B, Sage E (1999) Wavelength dependence of ultraviolet-induced DNA damage distribution: involvement of direct or indirect mechanisms and possible artefacts. J Photochem Photobiol B 49:71–80
Knebel A, Rahmsdorf HJ, Ullrich A, Herrlich P (1996) Dephosphorylation of receptor tyrosine kinases as target of regulation by radiation, oxidants or alkylating agents. EMBO J 15:5314–5325
Cavigelli M, Dolfi F, Claret FX, Karin M (1995) Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J 14:5957–5964
Tanos T, Marinissen MJ, Leskow FC, Hochbaum D, Martinetto H, Gutkind JS, Coso OA (2005) Phosphorylation of c-Fos by members of the p38 MAPK family. Role in the AP-1 response to UV light. J Biol Chem 280:18842–18852
Dunkern TR, Kaina B (2002) Cell proliferation and DNA breaks are involved in ultraviolet light-induced apoptosis in nucleotide excision repair-deficient Chinese hamster cells. Mol Biol Cell 13:348–361
Moorehead RA, Singh G (2000) Influence of the proto-oncogene c-fos on cisplatin sensitivity. Biochem Pharmacol 59:337–345
Saez E, Rutberg SE, Mueller E, Oppenheim H, Smoluk J, Yuspa SH, Spiegelman BM (1995) c-fos is required for malignant progression of skin tumors. Cell 82:721–732
Takahashi S, Pearse AD, Marks R (1994) Expression of c-fos proto-oncogene mRNA in non-melanoma skin cancer. J Dermatol Sci 7:54–62
Acknowledgments
We are grateful to L. Mullenders (Leiden, The Netherlands) for providing VH10tert cells. The work was supported by Deutsche Krebshilfe 106748, Deutsche Forschungsgemeinschaft CH 665/2-1 and Stiftung Rheinland Pfalz (no. 961-386261/809).
Conflict of interest
The authors declare that there is no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement 1.
Characterization of VH10tert cells(A) To analyze the p53 status, cells were exposed to 10 J/m2 UVC. At different times after exposure, cells were harvested, nuclear extracts were prepared, and 25 μg was subjected to Western blot analysis (c, non-exposed control). The filter was incubated with anti-p53, for loading control, with anti-ß-actin-specific antibodies. (B) Exponentially growing VH10tert cells were exposed to 10 J/m2 UVC. Total RNA was isolated at different time points after exposure, and real-time RT-PCR was performed using ddb2 and xpc or, as loading control, gapdh specific primers. (C) VH10tert cells were exposed to 10 or 25 J/m2. Cells were harvested at different time points after exposure, and genomic DNA was isolated. Equal amounts of DNA were blotted and incubated with anti-CPD (upper panel) or anti-(6-4)PP antibodies (lower panel) (con, non-exposed control). (D) VH10tert cells were exposed to different doses of UVC light; 96 h later cells were harvested and subjected to Sub-G1 measurement
Supplement 2. UVC-induced activation of the MAPK cascade and induction of Fos proteins in GM637 cells
(A) Exponentially growing GM637 cells were exposed to 10 J/m2 UVC. Total RNA was isolated at different time points after exposure, and semi-quantitative RT-PCR was performed using c-fos, fosB, fra1, or, as loading control, gapdh specific primers. (B/C) Exponentially growing GM637 cells were exposed to 10 J/m2 UVC for the indicated times. Protein extracts were prepared and subjected to Western blot analysis. The filter was incubated with c-Fos, FosB, or Fra1 specific antibodies (B) or incubated with p-JNK, JNK, p-p38 K, p38 K, p-ERK1/2 and ERK1/2 specific antibodies (C)
Supplement 3. Effect of c-Fos overexpression on
xpf/xpgexpression and proliferation (A) Human GM637 fibroblasts were transfected with a c-Fos expression plasmid. Parental GM637 cells and five clones expressing different amounts of c-Fos were selected, and the proliferation rate was determined. (B) Exponentially growing clones were exposed to 10 J/m2 UVC. Total RNA was isolated at different time points after exposure, and RT-PCR was performed using xpf, xpg, or, as positive control, gapdh specific primers
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Tomicic, M.T., Reischmann, P., Rasenberger, B. et al. Delayed c-Fos activation in human cells triggers XPF induction and an adaptive response to UVC-induced DNA damage and cytotoxicity. Cell. Mol. Life Sci. 68, 1785–1798 (2011). https://doi.org/10.1007/s00018-010-0546-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0546-9