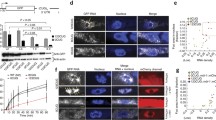
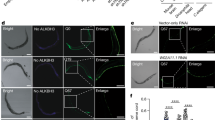
Abstract
We have utilized Caenorhabditis elegans as a model to investigate the toxicity and underlying mechanism of untranslated CAG repeats in comparison to CUG repeats. Our results indicate that CAG repeats can be toxic at the RNA level in a length-dependent manner, similar to that of CUG repeats. Both CAG and CUG repeats of toxic length form nuclear foci and co-localize with C. elegans muscleblind (CeMBL), implying that CeMBL may play a role in repeat RNA toxicity. Consistently, the phenotypes of worms expressing toxic CAG and CUG repeats, including shortened life span and reduced motility rate, were partially reversed by CeMbl over-expression. These results provide the first experimental evidence to show that the RNA toxicity induced by expanded CAG and CUG repeats can be mediated, at least in part, through the functional alteration of muscleblind in worms.







Similar content being viewed by others
References
Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, Day JW, Ranum LP (2006) Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet 38:758–769
Holmes SE, O’Hearn EE, McInnis MG, Gorelick-Feldman DA, Kleiderlein JJ, Callahan C, Kwak NG, Ingersoll-Ashworth RG, Sherr M, Sumner AJ, Sharp AH, Ananth U, Seltzer WK, Boss MA, Vieria-Saecker AM, Epplen JT, Riess O, Ross CA, Margolis RL (1999) Expansion of a novel CAG trinucleotide repeat in the 5′ region of PPP2R2B is associated with SCA12. Nat Genet 23:391–392
Holmes SE, O’Hearn E, Rosenblatt A, Callahan C, Hwang HS, Ingersoll-Ashworth RG, Fleisher A, Stevanin G, Brice A, Potter NT, Ross CA, Margolis RL (2001) A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat Genet 29:377–378
Ranum LP, Day JW (2004) Pathogenic RNA repeats: an expanding role in genetic disease. Trends Genet 20:506–512
Osborne RJ, Thornton CA (2006) RNA-dominant diseases. Hum Mol Genet 15(Spec No 2):R162–R169
Wheeler TM, Thornton CA (2007) Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol 20:572–576
Michlewski G, Krzyzosiak WJ (2005) Pathogenesis of spinocerebellar ataxias viewed from the RNA perspective. Cerebellum 4:19–24
Ranum LP, Cooper TA (2006) RNA-mediated neuromuscular disorders. Annu Rev Neurosci 29:259–277
Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA (2000) Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 289:1769–1773
Seznec H, Agbulut O, Sergeant N, Savouret C, Ghestem A, Tabti N, Willer JC, Ourth L, Duros C, Brisson E, Fouquet C, Butler-Browne G, Delacourte A, Junien C, Gourdon G (2001) Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum Mol Genet 10:2717–2726
Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS (1996) Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res 24:4407–4414
Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS (2000) Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J 19:4439–4448
Philips AV, Timchenko LT, Cooper TA (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280:737–741
Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS (2003) A muscleblind knockout model for myotonic dystrophy. Science 302:1978–1980
Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA (2004) Muscleblind proteins regulate alternative splicing. EMBO J 23:3103–3112
Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT (2001) RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem 276:7820–7826
Kuyumcu-Martinez NM, Wang GS, Cooper TA (2007) Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell 28:68–78
Ho TH, Bundman D, Armstrong DL, Cooper TA (2005) Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet 14:1539–1547
Begemann G, Paricio N, Artero R, Kiss I, Perez-Alonso M, Mlodzik M (1997) Muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development 124:4321–4331
Artero R, Prokop A, Paricio N, Begemann G, Pueyo I, Mlodzik M, Perez-Alonso M, Baylies MK (1998) The muscleblind gene participates in the organization of Z-bands and epidermal attachments of Drosophila muscles and is regulated by Dmef2. Dev Biol 195:131–143
Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD (2002) Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet 11:805–814
Squillace RM, Chenault DM, Wang EH (2002) Inhibition of muscle differentiation by the novel muscleblind-related protein CHCR. Dev Biol 250:218–230
Adereth Y, Dammai V, Kose N, Li R, Hsu T (2005) RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat Cell Biol 7:1240–1247
Lee KS, Smith K, Amieux PS, Wang EH (2008) MBNL3/CHCR prevents myogenic differentiation by inhibiting MyoD-dependent gene transcription. Differentiation 76:299–309
Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS (2006) Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA 103:11748–11753
Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP (2001) Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 293:864–867
Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA (2001) Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet 10:2165–2170
Savkur RS, Philips AV, Cooper TA, Dalton JC, Moseley ML, Ranum LP, Day JW (2004) Insulin receptor splicing alteration in myotonic dystrophy type 2. Am J Hum Genet 74:1309–1313
Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ (2003) RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res 31:5469–5482
Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA (2005) Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J Cell Sci 118:2923–2933
Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S (2004) Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum Mol Genet 13:495–507
Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS (2007) Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res 35:5474–5486
McLeod CJ, O’Keefe LV, Richards RI (2005) The pathogenic agent in Drosophila models of ‘polyglutamine’ diseases. Hum Mol Genet 14:1041–1048
Li LB, Yu Z, Teng X, Bonini NM (2008) RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 453:1107–1111
Faber PW, Alter JR, MacDonald ME, Hart AC (1999) Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci USA 96:179–184
Parker JA, Connolly JB, Wellington C, Hayden M, Dausset J, Neri C (2001) Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc Natl Acad Sci USA 98:13318–13323
Morley JF, Brignull HR, Weyers JJ, Morimoto RI (2002) The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA 99:10417–10422
Chen KY, Pan H, Lin MJ, Li YY, Wang LC, Wu YC, Hsiao KM (2007) Length-dependent toxicity of untranslated CUG repeats on Caenorhabditis elegans. Biochem Biophys Res Commun 352:774–779
Wang LC, Hung WT, Pan H, Chen KY, Wu YC, Liu YF, Hsiao KM (2008) Growth-dependent effect of muscleblind knockdown on Caenorhabditis elegans. Biochem Biophys Res Commun 366:705–709
Mello C, Fire A (1995) DNA transformation. Methods Cell Biol 48:451–482
Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77:71–94
Lewis JA, Fleming JT (1995) Basic culture methods. Methods Cell Biol 48:3–29
Waterston RH, Hirsh D, Lane TR (1984) Dominant mutations affecting muscle structure in Caenorhabditis elegans that map near the actin gene cluster. J Mol Biol 180:473–496
Hall DH (1995) Electron microscopy and 3D image reconstruction. In: Epstein HF, Shakes DC (eds) C. elegans: modern biological analysis of an organism. Methods in cell biology, vol. 48. Academic Press, New York, 395–436
Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, Meyer BJ (1997) Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci 17:8061–8073
Albertson DG, Fishpool RM, Birchall PS (1995) Fluorescence in situ hybridization for the detection of DNA and RNA. Methods Cell Biol 48:339–364
Fakhari FD, Dittmer DP (2002) Charting latency transcripts in Kaposi’s sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J Virol 76:6213–6223
Dittmer DP (2003) Transcription profile of Kaposi’s sarcoma-associated herpesvirus in primary Kaposi’s sarcoma lesions as determined by real-time PCR arrays. Cancer Res 63:2010–2015
Le Mee G, Ezzeddine N, Capri M, Ait-Ahmed O (2008) Repeat length and RNA expression level are not primary determinants in CUG expansion toxicity in Drosophila models. PLoS ONE 3:e1466
Galvao R, Mendes-Soares L, Camara J, Jaco I, Carmo-Fonseca M (2001) Triplet repeats, RNA secondary structure and toxic gain-of-function models for pathogenesis. Brain Res Bull 56:191–201
Goldberg YP, Kalchman MA, Metzler M, Nasir J, Zeisler J, Graham R, Koide HB, O’Kusky J, Sharp AH, Ross CA, Jirik F, Hayden MR (1996) Absence of disease phenotype and intergenerational stability of the CAG repeat in transgenic mice expressing the human Huntington disease transcript. Hum Mol Genet 5:177–185
Peel AL, Rao RV, Cottrell BA, Hayden MR, Ellerby LM, Bredesen DE (2001) Double-stranded RNA-dependent protein kinase, PKR, binds preferentially to Huntington’s disease (HD) transcripts and is activated in HD tissue. Hum Mol Genet 10:1531–1538
Klinkerfuss GH (1967) An electron microscopic study of myotonic dystrophy. Arch Neurol 16:181–193
Ludatscher RM, Kerner H, Amikam S, Gellei B (1978) Myotonia dystrophica with heart involvement: an electron microscopic study of skeletal, cardiac, and smooth muscle. J Clin Pathol 31:1057–1064
Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA (2000) Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA 6:79–87
Ebralidze A, Wang Y, Petkova V, Ebralidse K, Junghans RP (2004) RNA leaching of transcription factors disrupts transcription in myotonic dystrophy. Science 303:383–387
Osborne RJ, Lin X, Welle S, Sobczak K, O’Rourke JR, Swanson MS, Thornton CA (2009) Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet 18:1471–1481
Milne CA, Hodgkin J (1999) ETR-1, a homologue of a protein linked to myotonic dystrophy, is essential for muscle development in Caenorhabditis elegans. Curr Biol 9:1243–1246
Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, She WC, Luchak JM, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner PJ, McCall A, Canal I, Orr HT, Zoghbi HY, Botas J (2000) Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408:101–106
Siomi H, Dreyfuss G (1997) RNA-binding proteins as regulators of gene expression. Curr Opin Genet Dev 7:345–353
Acknowledgments
We are grateful to Dr. Y.-C. Wu (National Taiwan University) for providing myo-3::gfp and unc-54::mcherry plasmids and for discussion and reading the manuscript and to Dr. C.-S. Chen (National Cheng Kung University) for providing L4440::bre3 plasmid. This work was supported by the following grants from the National Science Council of Taiwan: NSC 95-2320-B-040-040-MY2 and NSC-97-2320-B-040-013-MY3 to H. Pan and NSC 96-2320-B-194-006-MY3 to K.-M. Hsiao.
Author information
Authors and Affiliations
Corresponding author
Additional information
L.-C. Wang, K.-Y. Chen and H. Pan contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.å
Suppl. Fig. 1
Muscle phenotype of CUG125 worms. (A) Electron microscopy images of GFP and CUG125 muscles. mcb, muscle cell body; M, M-line; db, dense body; bm, basement membrane; hyp, hypodermis; c, cuticle; Bar, 1 μm. (B) Dense body alignment in muscles of GFP and CUG125 worms. Anti-DEB-1 staining of dense bodies is shown as red spots. Arrow indicates the dense body within a muscle cell. Arrowhead indicates the dense body at the cell boundary. Bar, 10 μm (JPEG 327 kb)
Suppl. Fig. 2
Untranslated CAG and CUG repeats are not translated into polyQ protein. To investigate if CAG and CUG transgenes were translated into polyQ protein, Western blot analysis was performed using antibody against polyQ protein (Millipore, MAB1574) at a dilution of 1:3000 and total protein extracted from CAG83, CUG83 or Q78 transgenic worms. Q78 worms were generated by injection of myo3::gfp(Q78) plasmid (which contains CAG78 repeats in the coding region of gfp gene driven by the myo3 promoter) and were used as a positive control. 50 μg of total protein was loaded per well. The result indicates that untranslated CAG125 or CUG125 repeats in the transgenes do not produce a protein recognized by the antibody (JPEG 318 kb)
Suppl. Fig. 3
Reduced bre-3 gene expression by RNAi does not cause obvious effect on the life span of GFP, CUG83, and CAG83 worms. More than 30 transgenic worms at the F2 generation treated with bre-3(RNAi) or empty vector were assayed (JPEG 446 kb)
Suppl. Fig. 4
Reduced expression of deb-1a and deb-1c in CUG125, CAG125, and CeMbl(RNAi) worms. Semi-quantitative RT-PCR analyses of the deb-1a and deb-1c RNA level in GFP, CAG125 and CUG125 worms (left panel) or in control(RNAi) and CeMbl(RNAi) worms (right panel) were performed. Representative RT-PCR products were run on a 1% agarose gel and are shown in the figure. Actin served as an internal control for normalization (JPEG 285 kb)
Suppl. Fig. 5
The expression level of CeMbl was reduced in CAG125 worms. QPCR analysis of CeMbl RNA was performed using three cDNA preparations transcribed from one RNA sample. The RNA sample was collected from several CAG125 transgenic lines. In this experiment, the CeMbl RNA expression level in CAG125 worms is approximately 80% of that in GFP worms (P=0.001) (JPEG 190 kb)
Suppl. Fig. 6
Over-expression of CeMbl slightly increased the expression level of the gfp transcript containing CAG125 or CUG125 repeats. QPCR analysis of gfp transcript was performed using three cDNA preparations transcribed from one RNA sample. The RNA sample was collected from several CAG125 or CUG125 transgenic lines with or without co-expression of CeMbl-a. In this experiment, the average gfp RNA level in CUG125 and CAG125 worms co-expressing CeMbl is approximately 1.4 fold of that in control worms (JPEG 199 kb)
Suppl. Fig. 7
Either CeMbl-a or CeMbl-b over-expression can not reverse the (A) muscle morphology and (B) brood size of CUG125 and CAG125 worms. At least 30 worms of F3 to F4 generation from three independent injections were analyzed. Statistical analysis was performed using a student’s t-test. There is no significant difference between worms expressing untranslated CUG125 or CAG125 with or without CeMbl over-expression (JPEG 477 kb)
Rights and permissions
About this article
Cite this article
Wang, LC., Chen, KY., Pan, H. et al. Muscleblind participates in RNA toxicity of expanded CAG and CUG repeats in Caenorhabditis elegans . Cell. Mol. Life Sci. 68, 1255–1267 (2011). https://doi.org/10.1007/s00018-010-0522-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0522-4