Abstract
Myotonic dystrophy disorders are caused by expanded CUG repeats in noncoding regions. Here we used Caenorhabditis elegans expressing CUG repeats to identify genes that modulate the toxicity of such repeats. We identified 15 conserved genes that function as suppressors or enhancers of CUG repeat–induced toxicity and that modulate formation of nuclear foci by CUG-repeat RNA. These genes regulate CUG repeat–induced toxicity through distinct mechanisms including RNA export and clearance, thus suggesting that CUG-repeat toxicity is mediated by multiple pathways. A subset of the genes are also involved in other degenerative disorders. The nonsense-mediated mRNA decay (NMD) pathway has a conserved role in regulating CUG-repeat-RNA transcript levels and toxicity, and NMD recognition of toxic RNAs depends on 3′-untranslated-region GC-nucleotide content. Our studies suggest a broader surveillance role for NMD in which variations in this pathway influence multiple degenerative diseases.






Similar content being viewed by others
References
Todd, P.K. & Paulson, H.L. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 67, 291–300 (2010).
Brook, J.D. et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68, 799–808 (1992).
Taneja, K.L., McCurrach, M., Schalling, M., Housman, D. & Singer, R.H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 128, 995–1002 (1995).
Tokgozoglu, L.S. et al. Cardiac involvement in a large kindred with myotonic dystrophy: quantitative assessment and relation to size of CTG repeat expansion. J. Am. Med. Assoc. 274, 813–819 (1995).
Groh, W.J., Lowe, M.R., Simmons, Z., Bhakta, D. & Pascuzzi, R.M. Familial clustering of muscular and cardiac involvement in myotonic dystrophy type 1. Muscle Nerve 31, 719–724 (2005).
Miller, J.W. et al. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 19, 4439–4448 (2000).
Timchenko, N.A. et al. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 276, 7820–7826 (2001).
Yadava, R.S. et al. RNA toxicity in myotonic muscular dystrophy induces NKX2–5 expression. Nat. Genet. 40, 61–68 (2008).
Kim, D.H. et al. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 33, 3866–3874 (2005).
Garcia-Lopez, A. et al. Genetic and chemical modifiers of a CUG toxicity model in. Drosophila. PLoS ONE 3, e1595 (2008).
Osborne, R.J. et al. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum. Mol. Genet. 18, 1471–1481 (2009).
Du, H. et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 17, 187–193 (2010).
de Haro, M. et al. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum. Mol. Genet. 15, 2138–2145 (2006).
Mankodi, A. et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 289, 1769–1773 (2000).
Chen, K.Y. et al. Length-dependent toxicity of untranslated CUG repeats on Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 352, 774–779 (2007).
Raj, A., van den Bogaard, P., Rifkin, S.A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008).
Yuan, Y. et al. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 35, 5474–5486 (2007).
Jiang, H., Mankodi, A., Swanson, M.S., Moxley, R.T. & Thornton, C.A. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 13, 3079–3088 (2004).
Sasagawa, N., Ohno, E., Kino, Y., Watanabe, Y. & Ishiura, S. Identification of Caenorhabditis elegans K02H8.1 (CeMBL), a functional ortholog of mammalian MBNL proteins. J. Neurosci. Res. 87, 1090–1097 (2009).
Kim, J.K. et al. Functional genomic analysis of RNA interference in C. elegans. Science 308, 1164–1167 (2005).
Zhang, S., Binari, R., Zhou, R. & Perrimon, N. A genomewide RNA interference screen for modifiers of aggregates formation by mutant Huntingtin in Drosophila. Genetics 184, 1165–1179 (2010).
Illuzzi, J., Yerkes, S., Parekh-Olmedo, H. & Kmiec, E.B. DNA breakage and induction of DNA damage response proteins precede the appearance of visible mutant huntingtin aggregates. J. Neurosci. Res. 87, 733–747 (2009).
Bates, E.A., Victor, M., Jones, A.K., Shi, Y. & Hart, A.C. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J. Neurosci. 26, 2830–2838 (2006).
Ho, T.H. et al. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 118, 2923–2933 (2005).
Zhang, S., Ruiz-Echevarria, M.J., Quan, Y. & Peltz, S.W. Identification and characterization of a sequence motif involved in nonsense-mediated mRNA decay. Mol. Cell. Biol. 15, 2231–2244 (1995).
Jan, C.H., Friedman, R.C., Ruby, J.G. & Bartel, D.P. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469, 97–101 (2011).
Mangone, M. et al. The landscape of C. elegans 3′UTRs. Science 329, 432–435 (2010).
Zhang, C. et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 22, 2550–2563 (2008).
Li, L.B., Yu, Z., Teng, X. & Bonini, N.M. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 453, 1107–1111 (2008).
Nollen, E.A. et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA 101, 6403–6408 (2004).
Rehwinkel, J., Letunic, I., Raes, J., Bork, P. & Izaurralde, E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11, 1530–1544 (2005).
Wittmann, J., Hol, E.M. & Jack, H.M. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell. Biol. 26, 1272–1287 (2006).
Lemm, I. & Ross, J. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol. Cell. Biol. 22, 3959–3969 (2002).
Finkel, R.S. Read-through strategies for suppression of nonsense mutations in Duchenne/ Becker muscular dystrophy: aminoglycosides and ataluren (PTC124). J. Child Neurol. 25, 1158–1164 (2010).
Zetoune, A.B. et al. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 9, 83 (2008).
Resta, N. et al. A homozygous frameshift mutation in the ESCO2 gene: evidence of intertissue and interindividual variation in Nmd efficiency. J. Cell. Physiol. 209, 67–73 (2006).
Linde, L., Boelz, S., Neu-Yilik, G., Kulozik, A.E. & Kerem, B. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur. J. Hum. Genet. 15, 1156–1162 (2007).
Holbrook, J.A., Neu-Yilik, G., Hentze, M.W. & Kulozik, A.E. Nonsense-mediated decay approaches the clinic. Nat. Genet. 36, 801–808 (2004).
Amack, J.D. & Mahadevan, M.S. The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum. Mol. Genet. 10, 1879–1887 (2001).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Kamath, R.S., Martinez-Campos, M., Zipperlen, P., Fraser, A.G. & Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002 (2001).
Gidalevitz, T., Ben-Zvi, A., Ho, K.H., Brignull, H.R. & Morimoto, R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 (2006).
Montgomery, T.A. et al. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 8, e1002616 (2012).
Wu, X., Shi, Z., Cui, M., Han, M. & Ruvkun, G. Repression of germline RNAi pathways in somatic cells by retinoblastoma pathway chromatin complexes. PLoS Genet. 8, e1002542 (2012).
Acknowledgements
We thank M. Mahadevan (University of Virginia) for providing plasmids bearing the CUG repeats, S. Fischer and J. Kim for the RNAi library used in the screen, D. Kim (Massachusetts Institute of Technology) for the smg-2(qd101) strain and J. Lykke-Andersen (University of California, San Diego) for the mammalian antibody to UPF1. We are grateful to the J. Kaplan laboratory for the use of the Olympus FV-1000 confocal microscope and to the B. Seed lab for the use of their tissue-culture facilities. We are also thankful to S. Djonovic (Massachusetts General Hospital and Harvard Medical School) and S. Choi for reagents and technical assistance; J. Bai (Fred Hutchinson Cancer Research Center) for reagents, technical assistance and helpful discussions; A. Connery for help with the CellProfiler software; and J. Urbach for help editing the manuscript. We thank the Caenorhabditis elegans Genetics Center for strains; the American Heart Association (grant 0825938D) for funding S.M.D.A.G. and the US National Institutes of Health for funding G.R. (grant AG043184).
Author information
Authors and Affiliations
Contributions
S.M.D.A.G. and G.R. designed the study. S.M.D.A.G. performed the experiments and analyzed the data. Y.T. generated tools for SM-FISH image analysis and performed data analysis and statistical analysis. G.F.L. generated plasmids expressed in C. elegans and performed motility assays. M.A. set up the mammalian cell system and contributed to cell data analysis and interpretation. S.M.D.A.G. and G.R. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 C. elegans expressing expanded CUG repeats exhibit locomotion defects.
(A) Representation of motility assays performed using agar plates containing an E. coli food ring. The food ring had a 2 cm radius. (B) Motility assays for 2d adults. Data plotted corresponds to the average percentage of population to reach the food at each time point. Error bars represent SD from at least 3 independent experiments; in each experiment, 3-5 replicas of ca. 100-150 animals were analyzed. (C-E) Computational analysis of SM-FISH images. (C) Analysis starts with computational identification of the nuclear region based on DAPI staining in an SM-FISH image of a 123CUG animal. Following nucleus identification, (D) there is computational delineation of cytoplasmic versus nuclear spaces in the SM-FISH image corresponding to the GFP RNA transcript probes. (E) Analysis of pixel intensities for each SM-FISH image, corresponding to low RNA, high RNA densities and RNA foci in both the nucleus and cytoplasm.
Supplementary Figure 2 Expression of mbl-1::mCherry in C. elegans muscle cells increases expanded-CUG-transcript recruitment and nuclear-foci accumulation of mutant transcripts.
Schematic drawing of the MBL-1::mCHERRY construct (A), and C. elegans body wall muscle cells (B). (C) MBL-1::mCHERRY exhibits a diffuse cellular distribution with nuclear accumulation. (D) C. elegans muscle cells confocal SM-FISH images of GFP RNA transcripts (white), DAPI stained nucleus (blue), merge of GFP RNA and nucleus images, and mCherry translational fusion protein. The imaged muscle cells correspond to animals expressing 123CUG repeats and 0CUG, in a mCHERRY (control) or MBL-1::mCHERRY backgrounds. Yellow arrows indicate expanded CUG nuclear foci. MBL-1::mCHERRY localizes to the nucleus. (E) Genetic mosaic analysis of GFP intensity shows that GFP fluorescence, from 123CUG mRNA transcripts, absent in cells expressing mbl-1::mCherry, relative to neighboring cells that fail to express mbl-1::mCherry. GFP fluorescence is not affected in the 0CUG control animals expressing mbl-1::mCherry. (F) Confocal SM-FISH images of GFP RNA transcripts (white), DAPI stained nucleus (blue) and merge of C. elegans muscle cells. The strains imaged were 123CUG and 0CUG in: empty vector control (ctrl) and mbl-1 gene inactivations. Yellow arrows indicate expanded CUG nuclear foci. Bar, 1 μm.
Supplementary Figure 3 Screen approach for the identification of modulators of expanded-CUG toxicity.
(A) Representation of RNAi screen steps in the identification of modulators of expanded CUG repeat pathogenesis. (B) Fluorescent microscopy images of the strains 123CUG and the control 0CUG, on different RNAi gene inactivations: empty vector control (ctrl), mbl-1 and aly-3. Images were taken at the 3d old adult stage. Bar, 200 μm.
Supplementary Figure 4 Suppressors and enhancers of expanded-CUG toxicity have distinct effects on expanded-CUG nuclear-foci accumulation.
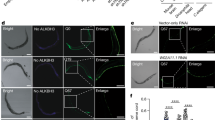
Confocal SM-FISH images of GFP RNA transcripts (white), DAPI stained nucleus (blue) and merge of C. elegans muscle cells. The strains imaged were 123CUG and the control 0CUG, in different RNAi gene inactivations: empty vector control (ctrl), C06A1.6, str-67, mrt-2, npp-4 and smg-2. Yellow arrows indicate expanded CUG nuclear foci.
Supplementary Figure 5 Gene inactivations have different effects on foci accumulation in the nucleus.
Computational analysis of SM-FISH images of 0CUG animals (yellow dots), control (green dots), and 123CUG animals fed different gene inactivations (blue dots) and control vector (red dots). Each ‘dot’ shown in the graph represents one analyzed SM-FISH image, corresponding to a single imaged cell. The red dotted square indicates the region of clustering of the samples corresponding to 123CUG animals on control vector (red dots). Labeled on the graph on the left, above the red box, are the gene inactivations (blue dots) that cause an increase in bright pixel intensity, corresponding to an increase in foci size or number, relative to the 123CUG on control. The ‘grouping’ of 123CUG npp-4 inactivations in the upper right corner of the graph indicates both an increase in nuclear foci and in nuclear ‘single’ transcript localization relative to the 0CUG npp-4 controls that localize further to the left in the graph. The inset section displayed shows gene inactivations (blue dots) that cause a decrease in bright pixel intensity, relative to the 123CUG on control vector, corresponding to a decrease in foci size or number. Axes correspond to fraction.
Supplementary Figure 6 Modulators of expanded-CUG foci accumulation in the nucleus
(A) Over-expression of expanded CUG repeat suppressors causes a decrease in expanded CUG nuclear foci accumulation. C. elegans muscle cells confocal SM-FISH images of GFP RNA transcripts (white), DAPI stained nucleus (blue), merge of GFP RNA and nucleus images, and mCherry translational fusion protein. The strains imaged are animals expressing 123CUG repeats and 0CUG in the following transgenic backgrounds: mCHERRY, NPP-4::mCHERRY, ASD-1::mCHERRY and RNP-2::mCHERRY. RNP-2 corresponds to the U1 small nuclear ribonucleoprotein A, and RNP-2::mCherry exhibits nuclear localization in C. elegans muscle cells. (B) Mutants in the NMD pathway cause an increase in expanded CUG nuclear foci accumulation. Confocal SM-FISH images of GFP RNA transcripts (white), DAPI stained nucleus (blue) and merge of C. elegans muscle cells. The strains imaged were animals expressing 123CUG repeats and 0CUG, in the following backgrounds: wild type (wt), smg-1(r861) and smg-6(r896). Yellow arrows indicate expanded CUG nuclear foci.
Supplementary Figure 7 NMD recognizes and degrades transcripts bearing GC-rich 3’ UTRs
(A-B) Sequence composition of CUG repeat sequences in the 3’UTR contributes to NMD transcript recognition for degradation. (A) Schematic drawing of the GC-rich or AT-rich plasmids for expressions in C. elegans muscle cells. (B) Fluorescent microscopy images of strains expressing a GFP with a 300bp ‘artificial’ insert in their 3’UTR containing the following GC percentages: 31%, 32%, 60% and 70%. Also included are the control strains containing 3’UTR inserts cloned from A. thaliana (34%GC) and P. aeruginosa (66%GC). These strains are shown in a wt background and in the background of the following smg mutants: smg-1(5861), smg-2(qd101) and smg-6(r896). The ‘fluorescence’ observed in the 60%GC and 70%GC strains in a wild type background corresponded to the characteristic gut autofluorescence, and no GFP signal was observed in the body wall muscle cells of these animals. Images were taken of animals at the L4 stage. Bar, 100 μm. (C-D) Western blot analysis of UPF1 down-regulation (24h post-transfection) by siRNA pool of unaffected (C) and DM1 (D) fibroblast cells, using UPF1-specific antibody. Fibroblasts showed a decrease of 40% in UPF1 levels relative to cells transfected with scrambled siRNAs (mock cells) in both unaffected (C) as well as DM1 (D) cells. GAPDH levels were used for normalization across samples.
Supplementary Figure 8 Model of regulation of expanded RNAs.
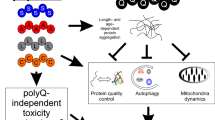
(A) Model for regulation of expanded RNA toxicity by the NMD pathway: NMD targets expanded CUG repeat transcripts for degradation reducing the levels of toxic RNAs present in the cells. A decrease in NMD function results in accumulation of toxic transcripts with increase in nuclear RNA foci and increase in toxicity with loss of motility. (B) Model for regulation of expanded RNA foci accumulation by the modulators of RNA toxicity identified: different pathways regulate expanded CUG repeat toxicity; an increase in foci causes a decrease in locomotion however, a decrease in foci doesn’t necessarily correlate with a decrease in muscle toxicity.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8, Supplementary Tables 1 and 2, and Supplementary Note (PDF 2468 kb)
Rights and permissions
About this article
Cite this article
Garcia, S., Tabach, Y., Lourenço, G. et al. Identification of genes in toxicity pathways of trinucleotide-repeat RNA in C. elegans . Nat Struct Mol Biol 21, 712–720 (2014). https://doi.org/10.1038/nsmb.2858
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2858
- Springer Nature America, Inc.
This article is cited by
-
Schlafen2 mutation in mice causes an osteopetrotic phenotype due to a decrease in the number of osteoclast progenitors
Scientific Reports (2018)
-
RNA biology of disease-associated microsatellite repeat expansions
Acta Neuropathologica Communications (2017)