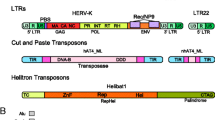
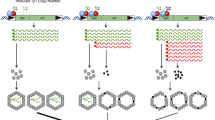
Abstract
Retroelements comprise a considerable fraction of eukaryotic genomes. Since their initial discovery by Barbara McClintock in maize DNA, retroelements have been found in genomes of almost all organisms. First considered as a “junk DNA” or genomic parasites, they were shown to influence genome functioning and to promote genetic innovations. For this reason, they were suggested as an important creative force in the genome evolution and adaptation of an organism to altered environmental conditions. In this review, we summarize the up-to-date knowledge of different ways of retroelement involvement in structural and functional evolution of genes and genomes, as well as the mechanisms generated by cells to control their retrotransposition.



Similar content being viewed by others
References
Bohne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN (2008) Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res 16:203–215
McClintock B (1956) Controlling elements and the gene. Cold Spring Harb Symp Quant Biol 21:197–216
Wessler SR (1998) Transposable elements and the evolution of gene expression. Symp Soc Exp Biol 51:115–122
Kapitonov VV, Jurka J (2003) Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci USA 100:6569–6574
International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Goodier JL, Kazazian HH Jr (2008) Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135:23–35
Beauregard A, Curcio MJ, Belfort M (2008) The take and give between retrotransposable elements and their hosts. Annu Rev Genet 42:587–617
Han JS, Boeke JD (2005) LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays 27:775–784
Bannert N, Kurth R (2004) Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA 101(suppl 2):14572–14579
Ohshima K, Hamada M, Terai Y, Okada N (1996) The 3′ ends of tRNA-derived short interspersed repetitive elements are derived from the 3′ ends of long interspersed repetitive elements. Mol Cell Biol 16:3756–3764
Evgen’ev MB, Arkhipova IR (2005) Penelope-like elements—a new class of retroelements: distribution, function and possible evolutionary significance. Cytogenet Genome Res 110:510–521
Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr (2003) Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA 100:5280–5285
Kazazian HH Jr, Moran JV (1998) The impact of L1 retrotransposons on the human genome. Nat Genet 19:19–24
Kazazian HH Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE (1988) Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332:164–166
Goodier JL, Zhang L, Vetter MR, Kazazian HH Jr (2007) LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol 27:6469–6483
Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr (1996) High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927
Luan DD, Korman MH, Jakubczak JL, Eickbush TH (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595–605
Feng Q, Moran JV, Kazazian HH Jr, Boeke JD (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905–916
Jurka J (1997) Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA 94:1872–1877
Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV (2001) Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol 21:1429–1439
Esnault C, Maestre J, Heidmann T (2000) Human LINE retrotransposons generate processed pseudogenes. Nat Genet 24:363–367
Gentles AJ, Wakefield MJ, Kohany O, Gu W, Batzer MA, Pollock DD, Jurka J (2007) Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res 17:992–1004
Donnelly SR, Hawkins TE, Moss SE (1999) A conserved nuclear element with a role in mammalian gene regulation. Hum Mol Genet 8:1723–1728
Eickbush TH (1992) Transposing without ends: the non-LTR retrotransposable elements. New Biol 4:430–440
Shen MR, Batzer MA, Deininger PL (1991) Evolution of the master Alu gene(s). J Mol Evol 33:311–320
Cordaux R, Hedges DJ, Herke SW, Batzer MA (2006) Estimating the retrotransposition rate of human Alu elements. Gene 373:134–137
Weiner AM, Deininger PL, Efstratiadis A (1986) Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem 55:631–661
Brosius J (1999) RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 238:115–134
Eickbush TH, Jamburuthugoda VK (2008) The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res 134:221–234
Leib-Mosch C, Seifarth W (1995) Evolution and biological significance of human retroelements. Virus Genes 11:133–145
Sverdlov ED (2000) Retroviruses and primate evolution. Bioessays 22:161–171
Mager D, Medstrand P (2002) Retroviral repeat sequences. In: Gardiner K (ed) Encyclopedia of the human genome. Nature Publishing, London
Poulter RT, Goodwin TJ (2005) DIRS-1 and the other tyrosine recombinase retrotransposons. Cytogenet Genome Res 110:575–588
Cappello J, Handelsman K, Lodish HF (1985) Sequence of Dictyostelium DIRS-1: an apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell 43:105–115
Goodwin TJ, Poulter RT (2004) A new group of tyrosine recombinase-encoding retrotransposons. Mol Biol Evol 21:746–759
Schostak N, Pyatkov K, Zelentsova E, Arkhipova I, Shagin D, Shagina I, Mudrik E, Blintsov A, Clark I, Finnegan DJ, Evgen’ev M (2008) Molecular dissection of Penelope transposable element regulatory machinery. Nucleic Acids Res 36:2522–2529
Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY (1994) Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon–intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem 269:8466–8476
Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA (2005) SVA elements: a hominid-specific retroposon family. J Mol Biol 354:994–1007
Ostertag EM, Goodier JL, Zhang Y, Kazazian HH Jr (2003) SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet 73:1444–1451
Malik HS, Eickbush TH (2001) Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res 11:1187–1197
Makalowski W (2000) Genomic scrap yard: how genomes utilize all that junk. Gene 259:61–67
Boissinot S, Davis J, Entezam A, Petrov D, Furano AV (2006) Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci USA 103:9590–9594
Song M, Boissinot S (2007) Selection against LINE-1 retrotransposons results principally from their ability to mediate ectopic recombination. Gene 390:206–213
Burwinkel B, Kilimann MW (1998) Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J Mol Biol 277:513–517
Segal Y, Peissel B, Renieri A, de Marchi M, Ballabio A, Pei Y, Zhou J (1999) LINE-1 elements at the sites of molecular rearrangements in Alport syndrome-diffuse leiomyomatosis. Am J Hum Genet 64:62–69
Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH (2000) Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet 9:2563–2572
Belancio VP, Hedges DJ, Deininger P (2008) Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res 18:343–358
Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, Jorde LB (2009) Mobile elements create structural variation: analysis of a complete human genome. Genome Res (in press)
Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, Dyer M, Cordaux R, Liang P, Batzer MA (2006) Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet 79:41–53
Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV (2002) DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet 31:159–165
Goodier JL, Ostertag EM, Kazazian HH Jr (2000) Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet 9:653–657
Pickeral OK, Makalowski W, Boguski MS, Boeke JD (2000) Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res 10:411–415
Moran JV, DeBerardinis RJ, Kazazian HH Jr (1999) Exon shuffling by L1 retrotransposition. Science 283:1530–1534
Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA (2006) Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci USA 103:17608–17613
Long M, Wang W, Zhang J (1999) Origin of new genes and source for N-terminal domain of the chimerical gene, jingwei, in Drosophila. Gene 238:135–141
Zaiss DM, Kloetzel PM (1999) A second gene encoding the mouse proteasome activator PA28beta subunit is part of a LINE1 element and is driven by a LINE1 promoter. J Mol Biol 287:829–835
Boschan C, Borchert A, Ufer C, Thiele BJ, Kuhn H (2002) Discovery of a functional retrotransposon of the murine phospholipid hydroperoxide glutathione peroxidase: chromosomal localization and tissue-specific expression pattern. Genomics 79:387–394
Babushok DV, Ostertag EM, Kazazian HH Jr (2007) Current topics in genome evolution: molecular mechanisms of new gene formation. Cell Mol Life Sci 64:542–554
Vinckenbosch N, Dupanloup I, Kaessmann H (2006) Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA 103:3220–3225
Temin HM (1993) Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA 90:6900–6903
Buzdin A, Gogvadze E, Lebrun MH (2007) Chimeric retrogenes suggest a role for the nucleolus in LINE amplification. FEBS Lett 581:2877–2882
Gogvadze E, Barbisan C, Lebrun MH, Buzdin A (2007) Tripartite chimeric pseudogene from the genome of rice blast fungus Magnaporthe grisea suggests double template jumps during long interspersed nuclear element (LINE) reverse transcription. BMC Genomics 8:360
Fudal I, Bohnert HU, Tharreau D, Lebrun MH (2005) Transposition of MINE, a composite retrotransposon, in the avirulence gene ACE1 of the rice blast fungus Magnaporthe grisea. Fungal Genet Biol 42:761–772
Buzdin A, Gogvadze E, Kovalskaya E, Volchkov P, Ustyugova S, Illarionova A, Fushan A, Vinogradova T, Sverdlov E (2003) The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res 31:4385–4390
Gilbert N, Lutz S, Morrish TA, Moran JV (2005) Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol 25:7780–7795
Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH Jr (2006) L1 integration in a transgenic mouse model. Genome Res 16:240–250
Nishihara H, Smit AF, Okada N (2006) Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res 16:864–874
Hayward BE, Zavanelli M, Furano AV (1997) Recombination creates novel L1 (LINE-1) elements in Rattus norvegicus. Genetics 146:641–654
Brosius J (1999) Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica 107:209–238
Furano AV (2000) The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol 64:255–294
van de Lagemaat LN, Landry JR, Mager DL, Medstrand P (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19:530–536
Polavarapu N, Marino-Ramirez L, Landsman D, McDonald JF, Jordan IK (2008) Evolutionary rates and patterns for human transcription factor binding sites derived from repetitive DNA. BMC Genomics 9:226
Landry JR, Rouhi A, Medstrand P, Mager DL (2002) The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol Biol Evol 19:1934–1942
Medstrand P, Landry JR, Mager DL (2001) Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J Biol Chem 276:1896–1903
Bieche I, Laurent A, Laurendeau I, Duret L, Giovangrandi Y, Frendo JL, Olivi M, Fausser JL, Evain-Brion D, Vidaud M (2003) Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol Reprod 68:1422–1429
Dunn CA, van de Lagemaat LN, Baillie GJ, Mager DL (2005) Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5. Gene 364:2–12
Romanish MT, Lock WM, de Lagemaat LN, Dunn CA, Mager DL (2007) Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet 3:e10
Carlton VE, Harris BZ, Puffenberger EG, Batta AK, Knisely AS, Robinson DL, Strauss KA, Shneider BL, Lim WA, Salen G, Morton DH, Bull LN (2003) Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet 34:91–96
Conley AB, Piriyapongsa J, Jordan IK (2008) Retroviral promoters in the human genome. Bioinformatics 24:1563–1567
Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E (2006) GREM, a technique for genome-wide isolation and quantitative analysis of promoter active repeats. Nucleic Acids Res 34:e67
Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E (2006) At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J Virol 80:10752–10762
Meisler MH, Ting CN (1993) The remarkable evolutionary history of the human amylase genes. Crit Rev Oral Biol Med 4:503–509
Long Q, Bengra C, Li C, Kutlar F, Tuan D (1998) A long terminal repeat of the human endogenous retrovirus ERV-9 is located in the 5′ boundary area of the human beta-globin locus control region. Genomics 54:542–555
Ling J, Pi W, Bollag R, Zeng S, Keskintepe M, Saliman H, Krantz S, Whitney B, Tuan D (2002) The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J Virol 76:2410–2423
Ling J, Pi W, Yu X, Bengra C, Long Q, Jin H, Seyfang A, Tuan D (2003) The ERV-9 LTR enhancer is not blocked by the HS5 insulator and synthesizes through the HS5 site non-coding, long RNAs that regulate LTR enhancer function. Nucleic Acids Res 31:4582–4596
Loreni F, Stavenhagen J, Kalff M, Robins DM (1988) A complex androgen-responsive enhancer resides 2 kilobases upstream of the mouse Slp gene. Mol Cell Biol 8:2350–2360
Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R (1998) Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem 273:891–897
Hambor JE, Mennone J, Coon ME, Hanke JH, Kavathas P (1993) Identification and characterization of an Alu-containing, T-cell-specific enhancer located in the last intron of the human CD8 alpha gene. Mol Cell Biol 13:7056–7070
Santangelo AM, de Souza FS, Franchini LF, Bumaschny VF, Low MJ, Rubinstein M (2007) Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet 3:1813–1826
Sasaki T, Nishihara H, Hirakawa M, Fujimura K, Tanaka M, Kokubo N, Kimura-Yoshida C, Matsuo I, Sumiyama K, Saitou N, Shimogori T, Okada N (2008) Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci USA 105:4220–4225
Corvelo A, Eyras E (2008) Exon creation and establishment in human genes. Genome Biol 9:R141
Lin L, Shen S, Tye A, Cai JJ, Jiang P, Davidson BL, Xing Y (2008) Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet 4:e1000225
Gotea V, Makalowski W (2006) Do transposable elements really contribute to proteomes? Trends Genet 22:260–267
Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G (2008) Intronic Alus influence alternative splicing. PLoS Genet 4:e1000204
Sorek R, Ast G, Graur D (2002) Alu-containing exons are alternatively spliced. Genome Res 12:1060–1067
Mola G, Vela E, Fernandez-Figueras MT, Isamat M, Munoz-Marmol AM (2007) Exonization of Alu-generated splice variants in the survivin gene of human and non-human primates. J Mol Biol 366:1055–1063
Gu Y, Kodama H, Watanabe S, Kikuchi N, Ishitsuka I, Ozawa H, Fujisawa C, Shiga K (2007) The first reported case of Menkes disease caused by an Alu insertion mutation. Brain Dev 29:105–108
Buzdin AA (2004) Retroelements and formation of chimeric retrogenes. Cell Mol Life Sci 61:2046–2059
Pavlicek A, Jabbari K, Paces J, Paces V, Hejnar JV, Bernardi G (2001) Similar integration but different stability of Alus and LINEs in the human genome. Gene 276:39–45
Belancio VP, Roy-Engel AM, Deininger P (2008) The impact of multiple splice sites in human L1 elements. Gene 411:38–45
Tamura M, Kajikawa M, Okada N (2007) Functional splice sites in a zebrafish LINE and their influence on zebrafish gene expression. Gene 390:221–231
Sironen A, Vilkki J, Bendixen C, Thomsen B (2007) Infertile Finnish Yorkshire boars carry a full-length LINE-1 retrotransposon within the KPL2 gene. Mol Genet Genomics 278:385–391
van de Lagemaat LN, Medstrand P, Mager DL (2006) Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol 7:R86
Hughes DC (2001) Alternative splicing of the human VEGFGR-3/FLT4 gene as a consequence of an integrated human endogenous retrovirus. J Mol Evol 53:77–79
Nishikura K (2006) Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol 7:919–931
Athanasiadis A, Rich A, Maas S (2004) Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2:e391
Borodulina OR, Kramerov DA (2008) Transcripts synthesized by RNA polymerase III can be polyadenylated in an AAUAAA-dependent manner. RNA 14:1865–1873
Cutter AD, Good JM, Pappas CT, Saunders MA, Starrett DM, Wheeler TJ (2005) Transposable element orientation bias in the Drosophila melanogaster genome. J Mol Evol 61:733–741
Baust C, Seifarth W, Germaier H, Hehlmann R, Leib-Mosch C (2000) HERV-K-T47D-related long terminal repeats mediate polyadenylation of cellular transcripts. Genomics 66:98–103
Kjellman C, Sjogren HO, Salford LG, Widegren B (1999) HERV-F (XA34) is a full-length human endogenous retrovirus expressed in placental and fetal tissues. Gene 239:99–107
Mager DL, Hunter DG, Schertzer M, Freeman JD (1999) Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3). Genomics 59:255–263
Han JS, Szak ST, Boeke JD (2004) Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429:268–274
Wheelan SJ, Aizawa Y, Han JS, Boeke JD (2005) Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res 15:1073–1078
Chen C, Ara T, Gautheret D (2009) Using Alu elements as polyadenylation sites: a case of retroposon exaptation. Mol Biol Evol 26:327–334
Lee JY, Ji Z, Tian B (2008) Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res 36:5581–5590
Trujillo MA, Sakagashira M, Eberhardt NL (2006) The human growth hormone gene contains a silencer embedded within an Alu repeat in the 3′-flanking region. Mol Endocrinol 20:2559–2575
Sharan C, Hamilton NM, Parl AK, Singh PK, Chaudhuri G (1999) Identification and characterization of a transcriptional silencer upstream of the human BRCA2 gene. Biochem Biophys Res Commun 265:285–290
Maeda N (1985) Nucleotide sequence of the haptoglobin and haptoglobin-related gene pair. The haptoglobin-related gene contains a retrovirus-like element. J Biol Chem 260:6698–6709
Hatada S, Grant DJ, Maeda N (2003) An intronic endogenous retrovirus-like sequence attenuates human haptoglobin-related gene expression in an orientation-dependent manner. Gene 319:55–63
Maeda N, Kim HS (1990) Three independent insertions of retrovirus-like sequences in the haptoglobin gene cluster of primates. Genomics 8:671–683
Medstrand P, van de Lagemaat LN, Mager DL (2002) Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res 12:1483–1495
Domansky AN, Kopantzev EP, Snezhkov EV, Lebedev YB, Leib-Mosch C, Sverdlov ED (2000) Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett 472:191–195
Dunn CA, Romanish MT, Gutierrez LE, van de Lagemaat LN, Mager DL (2006) Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene 366:335–342
Huh JW, Kim DS, Kang DW, Ha HS, Ahn K, Noh YN, Min DS, Chang KT, Kim HS (2008) Transcriptional regulation of GSDML gene by antisense-oriented HERV-H LTR element. Arch Virol 153:1201–1205
Matlik K, Redik K, Speek M (2006) L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol 2006:71753
Galante PA, Vidal DO, de Souza JE, Camargo AA, de Souza SJ (2007) Sense-antisense pairs in mammals: functional and evolutionary considerations. Genome Biol 8:R40
Conley AB, Miller WJ, Jordan IK (2008) Human cis natural antisense transcripts initiated by transposable elements. Trends Genet 24:53–56
Gogvadze E, Stukacheva E, Buzdin A, Sverdlov E (2009) Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J Virol 83:6098–6105
Kashkush K, Khasdan V (2007) Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes. Genetics 177:1975–1985
Smalheiser NR, Torvik VI (2005) Mammalian microRNAs derived from genomic repeats. Trends Genet 21:322–326
Hernandez-Pinzon I, de Jesus E, Santiago N, Casacuberta JM (2009) The frequent transcriptional readthrough of the tobacco tnt1 retrotransposon and its possible implications for the control of resistance genes. J Mol Evol 68:269–278
Piriyapongsa J, Jordan IK (2007) A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE 2:e203
Levy A, Sela N, Ast G (2008) TranspoGene and microTranspoGene: transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res 36:D47–D52
Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, Yung Y, Montoliu L, Glass CK, Rosenfeld MG (2007) Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 317:248–251
Akopov SB, Ruda VM, Batrak VV, Vetchinova AS, Chernov IP, Nikolaev LG, Bode J, Sverdlov ED (2006) Identification, genome mapping, and CTCF binding of potential insulators within the FXYD5-COX7A1 locus of human chromosome 19q13.12. Mamm Genome 17:1042–1049
Purbowasito W, Suda C, Yokomine T, Zubair M, Sado T, Tsutsui K, Sasaki H (2004) Large-scale identification and mapping of nuclear matrix-attachment regions in the distal imprinted domain of mouse chromosome 7. DNA Res 11:391–407
Dorsett D (1993) Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics 134:1135–1144
Girard L, Freeling M (1999) Regulatory changes as a consequence of transposon insertion. Dev Genet 25:291–296
Kostyuchenko MV, Savitskaya EE, Volkov IA, Golovnin AK, Georgiev PG (2008) Study of functional interaction between three copies of the insulator from the MDG4 transposable element in the model system of the miniwhite gene of Drosophila melanogaster. Dokl Biochem Biophys 421:239–243
Landry JR, Medstrand P, Mager DL (2001) Repetitive elements in the 5′ untranslated region of a human zinc-finger gene modulate transcription and translation efficiency. Genomics 76:110–116
Khanam T, Raabe CA, Kiefmann M, Handel S, Skryabin BV, Brosius J (2007) Can ID repetitive elements serve as cis-acting dendritic targeting elements? An in vivo study. PLoS ONE 2:e961
Chandler VL, Walbot V (1986) DNA modification of a maize transposable element correlates with loss of activity. Proc Natl Acad Sci USA 83:1767–1771
Weil C, Martienssen R (2008) Epigenetic interactions between transposons and genes: lessons from plants. Curr Opin Genet Dev 18:188–192
Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13:335–340
Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field JK, Liloglou T (2009) Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer 124:81–87
Arnaud P, Goubely C, Pelissier T, Deragon JM (2000) SINE retroposons can be used in vivo as nucleation centers for de novo methylation. Mol Cell Biol 20:3434–3441
Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM (1997) Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem 272:22322–22329
Lyon MF (2006) Do LINEs have a role in X-chromosome inactivation? J Biomed Biotechnol 2006:59746
Girard A, Hannon GJ (2008) Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 18:136–148
Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8:272–285
Klattenhoff C, Theurkauf W (2008) Biogenesis and germline functions of piRNAs. Development 135:3–9
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207
Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA (2007) Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res 35:5430–5438
Smalheiser NR, Torvik VI (2006) Alu elements within human mRNAs are probable microRNA targets. Trends Genet 22:532–536
Soifer HS, Zaragoza A, Peyvan M, Behlke MA, Rossi JJ (2005) A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res 33:846–856
Yang N, Kazazian HH Jr (2006) L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol 13:763–771
Schumann GG (2007) APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans 35:637–642
Acknowledgments
The authors were supported by the Molecular and Cellular Biology Program of the Presidium of the Russian Academy of Sciences, by the grant of the President of the Russian Federation and by the grant 08-04-00720-a and 09-04-12302 from the Russian Foundation for Basic Research. We apologize to authors whose primary references have not been cited due to space limitations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gogvadze, E., Buzdin, A. Retroelements and their impact on genome evolution and functioning. Cell. Mol. Life Sci. 66, 3727–3742 (2009). https://doi.org/10.1007/s00018-009-0107-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-009-0107-2