Abstract
Human intestinal organoids (HIOs) are a promising in vitro model consisting of different intestinal cell types with a 3D microarchitecture resembling native tissue. In the current study, we aimed to assess the expression of the most common intestinal CYP enzymes in a human induced pluripotent stem cell (hiPSC)-derived HIO model, and the suitability of that model to study chemical-induced changes in CYP expression and activity. We compared this model with the commonly used human colonic adenocarcinoma cell line Caco-2 and with a human primary intestinal epithelial cell (IEC)-based model, closely resembling in vivo tissue. We optimized an existing protocol to differentiate hiPSCs into HIOs and demonstrated that obtained HIOs contain a polarized epithelium with tight junctions consisting of enterocytes, goblet cells, enteroendocrine cells and Paneth cells. We extensively characterized the gene expression of CYPs and activity of CYP3A4/5, indicating relatively high gene expression levels of the most important intestinal CYP enzymes in HIOs compared to the other models. Furthermore, we showed that CYP1A1 and CYP1B1 were induced by β-naphtoflavone in all three models, whereas CYP3A4 was induced by phenobarbital and rifampicin in HIOs, in the IEC-based model (although not statistically significant), but not in Caco-2 cells. Interestingly, CYP2B6 expression was not induced in any of the models by the well-known liver CYP2B6 inducer phenobarbital. In conclusion, our study indicates that hiPSC-based HIOs are a useful in vitro intestinal model to study biotransformation of chemicals in the intestine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytochrome P450 (CYP) enzymes play crucial roles in detoxification or bioactivation of chemicals. Of the 18 CYP P450 families, members of the CYP 1–3 families are considered the main xenobiotic phase I metabolizing enzymes (Zanger and Schwab 2013). These enzymes are predominantly expressed in the liver, which is therefore the major organ for drug metabolism after oral exposure (Lin and Lu 2001; Paine et al. 2006). However, the intestine is regarded as the most important extrahepatic site of drug metabolism, especially upon oral exposure (Lin and Lu 2001; Paine et al. 2006). CYP enzymes are expressed in cells located at the tip of the villi and across the large surface area of the intestine, indicating that intestinal xenobiotic conversion can play an important role in overall first-pass metabolism (Murray et al. 1988; Xie et al. 2016).
In the small intestine, CYP3A4 has been observed to be the most abundant CYP enzyme. CYP2C9 and CYP3A5 have been reported to be the second most abundant enzymes, followed by CYP2C19, CYP2J2, CYP2D6 and CYP1A1 (Paine et al. 2006; Peters et al. 2016; Xie et al. 2016). However, large interindividual variations have been observed for, amongst others CYP3A5, CYP2D6 and CYP1A1, which were shown to be expressed in intestinal samples of some individuals, whereas they were absent in others (Paine et al. 2006; Peters et al. 2016; Xie et al. 2016). CYP expression and function is dependent on various factors, such as genetic polymorphisms, physiological factors (e.g., age, gender and (related) hormone levels) and pathological conditions (e.g., tumours and inflammation), which may impact chemical metabolism capacity (Cotreau et al. 2005; Zanger and Schwab 2013; Stavropoulou et al. 2018). Besides these factors, CYP functionality may also be affected by chemicals, e.g., by increasing their expression levels and/or inhibiting their activity, which may result in unanticipated adverse reactions and lower therapeutic efficacy of drugs (Lynch and Price 2007; Hansten 2018).
To date, human primary intestinal epithelial cells (IECs) are considered to be the best in vitro model to study intestinal metabolism (Grossmann et al. 2003), as well as primary IEC-derived models like the EpiIntestinal model (Ayehunie et al. 2018), as they represent the in vivo situation best. A disadvantage of human primary IECs is that they have been reported to survive for a limited time-frame, rapidly undergoing apoptosis when cultured ex vivo (Aldhous et al. 2001; Grossmann et al. 2003). This has been overcome by culturing the cells in a 3D-microtissue, like the human primary IEC-based EpiIntestinal model, which can be used for a longer time period. An alternative to the use of human primary cells is the use of cell lines, such as the human colonic adenocarcinoma cell line Caco-2, which is widely applied as a model to study intestinal transport, chemical metabolism and toxicity (Sun et al. 2008). Although Caco-2 cells form, upon differentiation, monolayers of polarized epithelial cells, which express various biotransformation enzymes, they generally show low biotransformation capacity and no (Prueksaritanont et al. 1996) or limited (Ozawa et al. 2015; Küblbeck et al. 2016) expression of CYP3A4, which is considered to play an important role in the intestinal metabolism of various chemicals, including midazolam and felodipine (Kato et al. 2003). This indicates that the Caco-2 model is limitedly suitable to study intestinal metabolism in vitro. Moreover, the Caco-2 model consists of a single cell type (enterocyte-like cells), and their typical culture in a 2D monolayer provides an in vitro intestinal tissue that lacks organ-specific microarchitecture and the physiological extracellular matrix environment (Eglen and Randle 2015).
Human intestinal organoids (HIOs) are promising novel in vitro models to study chemical metabolism, transport and toxicity. HIOs are 3D tissue structures with a microarchitecture and cellular composition resembling the native tissue (Spence et al. 2011; Finkbeiner et al. 2015; Tamminen et al. 2015). They can be generated by a stepwise differentiation process of human induced pluripotent stem cells (hiPSCs), by applying growth factors to mimic embryonic development (Sato et al. 2009, 2011; Mccracken et al. 2011; Finkbeiner et al. 2015). This differentiation process results in HIOs containing enterocytes, stem cells, goblet cells, Paneth cells, enteroendocrine cells and mesenchymal cells (Sato et al. 2009; Spence et al. 2011; Watson et al. 2014; Tamminen et al. 2015). The presence of mesenchymal cells enables crosstalk between epithelial and mesenchymal cells, which is known to have an essential role in in vivo intestinal development, for example in intestinal stem cell homeostasis (Kosinski et al. 2010; Le Guen et al. 2015; Meran et al. 2017). Therefore, hiPSC-derived HIOs are considered to be among the most promising in vitro models to assess chemical transport, metabolism and toxicity (Onozato et al. 2018). More specifically, these HIOs may overcome the limitations of the Caco-2 model regarding the limited expression of important biotransformation enzymes, such as CYP3A4.
In the current study we aimed to assess the suitability of hiPSC-derived HIOs as a model to study chemical-induced changes in CYP-expression and CYP activity, by characterizing the gene expression of the most common intestinal CYP enzymes and their inducibility in hiPSC-derived HIOs, as well as gene expression of some common intestinal enzymes involved in phase II metabolism. Caco-2 cells were used for comparison throughout all experiments and the EpiIntestinal model was used for comparison of the gene expression studies. HIOs were obtained based on differentiation methods described by McCracken et al. (2011), Spence et al. (2011) and Onozato et al. (2018), which were further optimized in the present study. HIOs were extensively characterized during the various differentiation stages and the optimisation of the differentiation method was supported by characterization of CYP and differentiation marker gene expression profiles and by immunohistochemical analysis. Using the optimized protocol, CYP expression was further evaluated in the absence or presence of typical CYP inducers (β-naphthoflavone, phenobarbital and rifampicin) and compared with expression in Caco-2 cells and the EpiIntestinal model. Induction of CYP3A4/5 activity was assessed by measuring midazolam hydroxylation (1′-OH-midazolam and 4-OH-midazolam formation) in HIOs and Caco-2 cells. The present study provides insight into whether HIOs are a relevant in vitro intestinal model regarding chemical metabolism and chemical-induced changes in expression and activity of biotransformation enzymes.
Materials and methods
Caco-2 cell culture
Human colorectal adenocarcinoma cells (Caco-2, HTB-37) were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cells were cultured in a humidified incubator (37 °C, 5% CO2) in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Thermo Fisher Scientific, Waltham, MA), 1% non-essential amino acids (Gibco) and 1% penicillin/streptomycin (Sigma, St. Louis, MO). For experiments, Caco-2 cells were harvested at 80% confluence and seeded at a density of 4 × 105 cells/cm2 into inserts with a polycarbonate membrane (0.4 µm pore, Corning, New York, NY). After 21 days of cultivation (using 500 µl in the apical and 1500 µl in the basolateral compartment), monolayer integrity was determined by measuring transepithelial electrical resistance (TEER) with a Millicell-ERS Volt-Ohm meter (Millipore). Only monolayers exhibiting TEER values exceeding 400 Ω cm2 were used for experiments.
EpiIntestinal culture
The EpiIntestinal 3D tissue model of the human small intestine (MatTek Life Sciences, Ashland, USA) consists of enterocytes, Paneth cells, M cells, tuft cells and intestinal stem cells cultured in permeable cell culture inserts. Cell culture inserts (8.8 mm ID) were placed in 12-well plates and cells were maintained in 100 µl maintenance medium (MatTek Life Sciences) in the apical compartment and 5 ml maintenance medium in the basolateral compartment upon arrival. For CYP induction studies, maintenance medium was replaced with maintenance medium with CYP inducers 1 day after arrival.
hiPSC culture
The hiPSC cell line (CS83iCTR-33n1) was provided by the Cedars-Sinai Medical Center’s David and Janet Polak Foundation Stem Cell Core Laboratory. These cells have been generated through episomal reprogramming of fibroblasts of a 31-year-old healthy female. The cell line has been fully characterized and no karyotype abnormalities have been found. hiPSCs were cultured on growth factor-reduced matrigel-coated (Corning) cell culture plates in mTeSR1 medium (Stem Cell Technologies, Vancouver, Canada) and were passaged using gentle cell dissociation reagent (Stem Cell Technologies).
hiPSC differentiation into intestinal organoids
For differentiation, hiPSCs were dissociated into single cells using accutase (Stem Cell Technologies) and cultured on human embryonic stem cell qualified matrigel-coated 24-well plates in mTeSR1 supplemented with 10 µM Y-27632 (Stem Cell Technologies) for 1 day. hiPSCs were subsequently differentiated into definitive endoderm (DE) by incubation in RPMI1640 medium containing 1% non-essential amino acids (Gibco), 100 ng/ml Activin A (Cell Guidance Systems, Cambridge, UK) with increasing concentrations of fetal bovine serum (0%, 0.2% and 2% on day 1, 2 and 3, respectively). 15 ng/ml BMP4 (R&D Systems, Minneapolis, MN) was also added during the first day of definitive endoderm formation. Hindgut endoderm formation was induced by changing the medium to RPMI1640 medium containing 1% non-essential amino acids (Gibco), 2% fetal bovine serum, 500 ng/ml FGF4 (R&D Systems) and 3 µM Chiron99021 (Stemgent). After 4 days, free-floating spheroids were collected and embedded into domes of Matrigel (Corning). Spheroids were cultured in Advanced DMEM/F-12 containing 1 × B27 (Gibco), 1 × N2 (Gibco), 15 mM HEPES (Gibco), 1% penicillin/streptomycin (Gibco), 2 mM l-glutamine (Gibco), 50 ng/ml EGF (R&D Systems), 100 ng/ml Noggin (R&D Systems) and 500 ng/ml RSpondin-1 (R&D Systems). Medium was refreshed every 2–3 days and HIOs were passaged every 10–14 days. Organoids used for experiments where grown in Matrigel for at least 28 days unless otherwise stated. To promote intestinal differentiation, 0.5 µM A-83-01 (Sigma), 20 µM PD98059 (Stem Cell Technologies), 5 mM 5-aza-2′-deoxycytidine (Sigma), and 5 µM DAPT (Sigma) were added to the medium either from day 14 to 28 (according to Onozato et al. 2018), or after day 28 for a period of 3 or 7 days (further optimized protocol present study).
RNA isolation and qPCR
Total RNA was extracted from the Caco-2 cells using the RNeasy Mini Kit (Qiagen, Venlo, The Netherlands). Total RNA was extracted from hiPSCs, organoids and EpiIntestinal tissues using the NucleoSpin RNA isolation kit (Macherey-Nagel, Düren, Germany). Subsequently, 200–500 ng RNA was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Veenendaal, The Netherlands). Changes in gene expression were determined by real-time PCR on a CFX384 real-time PCR detection system (Bio-Rad Laboratories) using SensiMix (Bioline; GC Biotech, Alphen aan den Rijn, The Netherlands). The PCR conditions consisted of an initial denaturation of 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing extension at 60 °C for 15 s. The housekeeping gene RPL27 was used for normalization of gene expression when assessing the influence of time of differentiation on gene expression (HIOs), the gene expression of CYP enzymes when comparing culture protocols (HIOs) and the gene expression of CYP enzymes after chemical treatment (EpiIntestinal model, HIOs, Caco-2 cells). For comparison of gene expression of CYP and other selected phase I and phase II biotranformation enzymes between Caco-2 cells, HIOs and the EpiIntestinal model without chemical treatment, gene expression levels were normalized on Ct levels, since the expression of RPL27 was different in the different cell models. Primer sequences were taken from the Harvard PrimerBank and ordered from Eurogentec (Liège, Belgium). Sequences of the used primers are listed in Table 1.
Immunofluorescence staining
Intestinal organoids were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA), transferred to a 30% sucrose solution overnight at 4 °C, frozen in Tissue-Tek O.C.T. compound (Sakura Finetek Europe, Alphen aan den Rijn, the Netherlands) and cut in 10 µm sections. The sections were subsequently permeabilized in 0.5% Triton X-100 (Sigma) in Phosphate Buffered Saline (PBS, Gibco) and blocked using 5% normal donkey serum (Jackson ImmunoResearch, West Grove, PA) in 0.5% Triton X-100 in PBS. Primary antibodies were applied overnight at 4 °C in blocking buffer. The next day, sections were washed and incubated with appropriate secondary antibody for 1 h at room temperature. The antibodies including their dilutions are listed in Table 2. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen). Finally, sections were washed and mounted in ProLong Diamond Antifade Mountant (Invitrogen). Confocal images were captured on a Zeiss LSM510 microscope.
Induction of CYP gene expression
For these studies, 24 h prior to exposure, three HIOs were pooled in one Matrigel dome. HIOs, the EpiIntestinal model and Caco-2 cells were exposed to 50 µM β-naphthoflavone (Sigma), 500 µM phenobarbital (Bipharma, Almere, the Netherlands) or 50 µM rifampicin (Supelco, Bellefonte, PA) for 48 h. Stock solutions were made in DMSO (Sigma) and final concentrations amounted to 0.1% in the medium. HIOs were exposed to 500 µl exposure medium. Caco-2 cells were exposed to 500 µl exposure medium in the apical and 1500 µl exposure medium in the basolateral compartment, and EpiIntestinal tissues were exposed to 50 µl exposure medium in the apical and 5 ml exposure medium in the basolateral compartment. Exposure medium was refreshed after 24 h. After incubation, HIOs, Caco-2 cells and the EpiIntestinal tissues were washed with PBS, collected and stored at − 80 °C before RNA isolation.
Induction of CYP3A4/5 activity
CYP3A4/5 activity and its induction by rifampicin was determined by measuring the formation of 1′-hydroxymidazolam and 4-hydroxymidazolam in non-exposed and rifampicin-exposed HIOs and Caco-2 cells. For these studies, 24 h prior to exposure, three HIOs were pooled in one Matrigel dome. HIOs were exposed to 50 µM rifampicin for 48 h (final DMSO concentration: 0.1%). Subsequently, HIOs were removed from the Matrigel and each organoid was cut into four pieces. These organoids were transferred to 96-well ultra-low attachment microplates (Corning) and incubated in 200 µl Advanced DMEM/F-12 containing 1 × B27, 1 × N2, 15 mM HEPES, 1% penicillin/streptomycin, 2 mM l-glutamine, 50 ng/ml EGF (R&D Systems), 100 ng/ml Noggin (R&D Systems) and 500 ng/ml RSpondin-1 (R&D Systems) for 1 h. Subsequently, medium was replaced with fresh medium containing 50 µM midazolam (European Pharmacopoeia Reference, Strasbourg, France). After 24 h, 100 µl medium was collected and 100 µl ice-cold acetonitrile (Actu-All Chemicals, Oss, the Netherlands) was added to precipitate the proteins in the medium. The organoids were washed five times with PBS and organoid homogenates were prepared by lysing organoids in 250 µl ice-cold RIPA buffer (Pierce, Thermo Fisher Scientific). After centrifugation, 100 µl organoid lysate was collected and 100 µl acetonitrile was added to precipitate the proteins. The remainder of the cell lysate was used for protein quantification using the BCA Protein Assay kit (Pierce, Thermo Fisher Scientific) to normalize CYP3A/5 activity to protein levels.
After 21 days of differentiation, Caco-2 cells were exposed to 50 µM rifampicin for 48 h (final DMSO concentration: 0.1%). Culture medium was subsequently replaced with fresh DMEM containing 10% heat-inactivated fetal bovine serum, 1% non-essential amino acids, 1% penicillin/streptomycin and 50 µM midazolam (final DMSO concentration: 0.1%). After 24 h incubation, 100 µl medium was collected, (25 µl from the apical compartment and 75 µl from the basolateral compartments, reflecting the ratio of total volumes in the apical (500 µl) and basolateral (1500 µl) compartments), and 100 µl ice-cold acetonitrile was added to precipitate the proteins. The Caco-2 cells were washed five times with PBS and cell lysates were prepared by lysing Caco-2 cells in 250 µl ice-cold RIPA buffer (Pierce, Thermo Fisher Scientific). After centrifugation, 100 µl Caco-2 cell lysate was collected and 100 µl acetonitrile was added to precipitate the proteins. The remainder of the cell lysate was used for protein quantification using the BCA Protein Assay kit (Pierce, Thermo Fisher Scientific) to normalize CYP3A4/5 activity to protein levels.
LC–MS analysis
The formation of 1′-OH-midazolam and 4-OH-midazolam was quantified in the culture medium and cell lysates of Caco-2 cells and HIOs using an Ultimate 3000 UHPLC system coupled to a Q-Exactive Orbitrap™-based mass spectrometer with a HESI-II electrospray operating in positive ion mode, using the vDIA method as described previously (Zomer and Mol 2015). The eluents for the LC separation were (A) water and (B) methanol:water 95:5 (v/v) both containing 2 mM ammonium formate and 20 µl formic acid per litre. The following gradient was used: 0% B until 0.1 min, then linear to 45% B in 1.9 min, followed by a rise to 100% B in 6 min. This condition was held for 6 min, after which a switch back to 0% B was performed in 0.5 min. After 4.5 min of equilibration, the next sample was injected. An Atlantis T3 LC column (3 µm particles, 100 × 3 mm) (Waters, Milford, MA, USA) was used. The LC flow rate was 300 µl/min, the LC column was kept at 40 °C and the injection volume was 5 µl. Under these conditions, retention times for 1′-OH-midazolam and 4-OH-midazolam were 10.1 and 9.8 min, respectively. Commercially available 1′-OH-midazolam (LGC Standards GmbH, Wesel, Germany) and 4-OH-midazolam (Sigma) were used to prepare standard curves. For quantification of 1′-OH-midazolam and 4-OH-midazolam, the protonated molecule (m/z: 342.08039) was used with a mass tolerance of ± 5 ppm. Additionally, fragments were used for identification of 1′-OH-midazolam (C11H8N2+ m/z: 168.06819) and 4-OH-midazolam (C13H10ClNF+ m/z: 234.0480). The software package Tracefinder (version 4.1, Thermo Scientific) was used to process the data.
Statistical analysis
Data are presented as mean ± SEM. Comparisons between hiPSC-derived organoids and Caco-2 cells and between hiPSC-derived organoids with and without added small compounds were analysed using a two-tailed Student’s t test. A one-way ANOVA followed by Tukey’s post hoc multiple comparison test was used for comparisons between hiPSC-derived organoids, Caco-2 cells and the EpiIntestinal tissues and to determine statistically significant differences in the CYP induction experiments. P < 0.05 was considered as statistically significant. Prism software (version 5.02; GraphPad, San Diego, CA) was used for statistical analysis.
Results
Differentiation and characterization of intestinal organoids
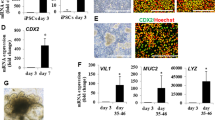
The differentiation procedure of the human iPSC line CS83iCTR-33n1 into intestinal organoids was based on previous reports (Mccracken et al. 2011; Spence et al. 2011) (Fig. 1a). In the present study, the differentiation procedure was characterized by gene expression measurements and immunohistochemical evaluation. hiPSCs were first differentiated into definitive endoderm (DE) using the nodal-mimetic Activin A and BMP4. The provision of BMP4 to Activin A at the onset of differentiation promotes DE formation (Teo et al. 2012). Although mRNA levels of the pluripotency markers OCT4 and NANOG were not altered in the generated DE cells (Fig. 1b), robust induction of the DE markers FOXA2, SOX17, CXCR4 and GSC was observed (Fig. 1c). The DE cells were subsequently treated with FGF4 and Chiron99021 to induce hindgut endoderm formation and intestinal specification. Chiron99021 very potently inhibits the glycogen synthase kinase 3 (GSK3) pathway, resulting in the activation of WNT signalling and has been shown to be more potent in inducing hindgut endoderm formation than the more commonly used WNT3A (Tamminen et al. 2015). During the 4 days exposure to FGF4 and Chiron99021, the flat sheet of DE cells transformed into a hindgut endoderm culture, characterized by the expression of Caudal-related homeobox 2 (CDX2) (Fig. 1d), that started budding off and formed free-floating hindgut spheroids (Fig. 1e). Importantly, mRNA levels of pluripotency and definitive endoderm markers were lower in the hindgut endoderm culture and hindgut spheroids compared to the definitive endoderm stage (Fig. 1b, c). The spheroids were subsequently embedded into Matrigel (HIO 0 days) to promote intestinal differentiation and growth (Fig. 1g). After 14, 28 and 42 days in 3D culture, HIOs were harvested to determine gene expression levels of typical intestinal markers. As compared to spheroids, gene expression levels of intestinal differentiation markers KRT20 (cellular protein specific for mature enterocytes and goblet cells), KLF5 (transcription factor important for maintenance of barrier integrity and crypt architecture), IFABP2 (cellular protein specific for mature enterocytes) and VIL1 (major component brush border cytoskeleton specific for enterocytes) gradually increased over time in the developing HIOs (Fig. 1d). Although expression of the intestinal transcription factor CDX2 was lower in HIOs in 3D culture than in spheroids (Fig. 1d), CDX2 was still clearly detected in virtually all epithelial cells (Fig. 1h). The intestinal crypt/stem cell markers SOX9 and LGR5 were both well expressed in the spheroids and expression gradually increased over time up to 42 days in HIO 3D culture. Interestingly, LGR5 expression already peaked in the DE phase (Fig. 1d). In addition to enterocytes, the HIOs also contain Paneth cells, goblet cells and enteroendocrine cells as evidenced by expression of LYZ, MUC2 and CHGA, respectively (Fig. 1f). Although CHGA expression peaked in HIOs that were 14 days in Matrigel, CHGA was still well expressed in HIOs that were 28 and 42 days in 3D culture, as evidenced by an average Ct value of 25 and by immunofluorescence staining showing the presence of CHGA + cells in the HIOs (Fig. 1f, h). Goblet cells (MUC2 +) were also identified microscopically in the epithelium of the HIOs (Fig. 1h). The presence of E-cadherin, ZO-1 and Villin1 located towards the apical surface of the enterocytes, demonstrates that the epithelial cells are polarized and formed tight junctions (Fig. 1h). Importantly, the increase in gene expression of FOXF1, VIM and ACTA2 during HIO formation and the presence of Vimentin + cells indicated that the HIOs also contained mesenchymal cells (Fig. 1f, h). Although HIOs that were in 3D culture for 42 days obtained highest gene expression levels for the majority of the intestinal differentiation markers, genes related to the four major intestinal cell types (enterocytes, goblet cells, enteroendocrine and Paneth cells) were already well expressed after 28 days. Therefore, HIOs were kept at least 28 days in 3D culture before performing experiments, which is in line with McCracken et al. (2011) and Spence et al. (2011).
Differentiation of hiPSCs into human intestinal organoids (HIOs). a Schematic procedure for the differentiation of the hiPSC line CS83iCTR-33n1 into HIOs. Relative expression of b pluripotency and c definitive endoderm (DE) markers during the course of differentiation up to HIOs that were 42 days in 3D culture. The highest expression levels of each gene in each differentiation phase were set at one. HG, hindgut endoderm. d Relative expression of intestinal differentiation and crypt/stem cell markers during the course of differentiation up to HIOs that were 42 days in 3D culture. The highest expression levels of each gene in each differentiation phase were set at one. e Microscopic images of hiPSC-derived spheroids on day 7 of the differentiation procedure. f Relative expression of markers of Paneth cells (LYZ), goblet cells (MUC2), enteroendocrine cells (CHGA) and mesenchymal cells (FOXF1, VIM and ACTA2) during the course of differentiation up to HIOs that were 42 days in 3D culture. The highest expression levels of each gene in each differentiation phase were set at one. g Microscopic images of HIOs in 3D culture for 28 or 42 days. h Immunofluorescence stainings of various intestinal differentiation markers in HIOs that were in 3D culture for at least 28 days. Nuclei were counterstained with 4′,d-diamidino-2-phenylindole (DAPI). E-CAD: E-cadherin, CHGA: chromogranin A, ZO-1: zonula occludens-1, MUC2: mucin 2, VIM: vimentin. Data are mean values ± SEM from triplicate wells
CYP gene expression
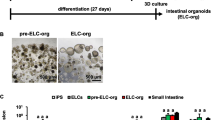
We determined the gene expression of a series of relevant intestinal CYP enzymes in HIOs and compared these with expression levels in Caco-2 cells. Whereas gene expression of CYP1B1, CYP2B6, CYP2C9 and CYP3A5 was significantly higher in HIOs, gene expression of CYP1A1 and CYP2C19 was significantly lower in the HIOs as compared to Caco-2 cells (Fig. 2a, Supplemental Table 1). Of note, both HIOs and Caco-2 cells did not express CYP2D6 (data not shown), a gene containing the highest number of allele variants amongst the CYP enzymes resulting in CYP2D6 gene deletion in approximately 5% of Asians, Africans and Caucasians (Ingelman-Sundberg et al. 2007). CYP3A4, with a Ct value of 32, showed limited expression in both Caco-2 cells and HIOs (Fig. 2a, Supplemental Table 1). Recently, Onozato et al. (2018) reported that small-molecule compounds (A-83-01, PD98059, 5-aza-2′-deoxycytidine and DAPT) induce expression of various genes in hiPSC-derived HIOs, including CYP3A4, when added to the medium during the final 15 days of the differentiation period [starting from HIOs of 12 days (differentiation day 19)]. Therefore, we also differentiated hiPSCs into HIOs in the presence or absence of small-molecule compounds and assessed the expression of the CYP genes. Adding small-molecule compounds during HIO differentiation from day 21 to 35 resulted in mRNA levels of CYP3A4 that were about 900-fold higher as compared to HIOs differentiated in the absence of small-molecule compounds (Fig. 2b, Supplemental Table 2). In addition, small-molecule compounds robustly induced the expression of other CYP enzymes, including CYP1A1, CYP1B1, CYP2C9, CYP2C19, CYP2J2 and CYP3A5, but not CYP2B6 (Fig. 2b, Supplemental Table 2). A disadvantage of adding small-molecule compounds during HIO differentiation was that HIO growth almost completely stopped, indicating that the organoid culture cannot be further expanded and one would need to start differentiation from the hiPSCs if more organoids are needed. Also, the HIOs displayed a more compact morphology (Fig. 2c). Thus, we aimed to optimise the differentiation protocol into a protocol that can be used to keep a culture of well-growing HIOs, using the protocol as depicted in Fig. 1a, and supply fully maturated HIOs by treating a subset of the HIOs with small molecule compounds. To that end, HIOs were allowed to differentiate and grow for at least 35 days (28-day old HIOs) and were subsequently treated with small-molecule compounds for either 3 or 7 days. Figure 2d and e (Supplemental Tables 3 and 4) show that the relative mRNA levels in small molecule compounds-treated HIOs were similar for most CYPs when using a 7-day treatment with small-molecule compounds as when using the protocol of Onozato et al. (2018) (Fig. 2b, e), whereas these were lower when using a 3-day treatment with small-molecule compounds (Fig. 2d). For example, relative CYP3A4 expression levels were about 60-fold higher than the control after 3 days incubation with small-molecule compounds, whereas 7 days incubation resulted in an 800-fold increase versus the control (Fig. 2d, e), being almost the same as the 900-fold increase versus the control in relative CYP3A4 expression when using the protocol of Onozato et al. (2018) (Fig. 2b).
CYP gene expression in Caco-2 cells and HIOs. a Relative gene expression of common intestinal CYP enzymes in Caco-2 cells and HIOs. b Schematic procedure for HIO formation in the presence of the small-molecule compounds (A, A-83-01; PD, PD98059; 5-AZA, 5-aza-2′-deoxycytidine; DAPT) from day 21 to 35. Relative gene expression of common intestinal CYP enzymes in HIOs differentiated in the absence or presence of small-molecule compounds for 14 days. Gene expression levels in HIOs differentiated in the absence of small-molecule compounds (CTRL) were set at one. c Microscopic images of HIOs differentiated in the absence (CTRL) or presence of small-molecule compounds for 14 days. Schematic procedure for HIOs exposed to small-molecule compounds for d 3 or e 7 days after the organoid differentiation phase (after day 35) and relative gene expression of common intestinal CYP enzymes in HIOs exposed to small-molecule compounds. Gene expression levels in HIOs not exposed to small-molecule compounds (CTRL) were set at one. f CYP3A4/5 activity, quantified by 1′-OH-midazolam (1′-OH-MDZ) and 4-OH-midazolam (4-OH-MDZ) formation in HIO lysates and medium, in HIOs exposed to small-molecule compounds for 7 days. CYP3A4/5 activity in HIOs not exposed to small-molecule compounds (CTRL) are also presented. Asterisks indicate significantly different compared with Caco-2 cells or CTRL. *P < 0.05, **P < 0.001. Data are mean values ± SEM from triplicate wells
Interestingly, 7 days provision of small-molecule compounds also promoted HIO differentiation as displayed by elevated gene expression levels of intestinal differentiation markers, and markers for goblet cells and enteroendocrine cells (Supplemental Fig. 1a, b). These HIOs also contained a polarized epithelium with tight junctions and all different cell types (Supplemental Fig. 1c).
Activity of CYP3A4/5 in HIOs treated with small-molecule compounds for 7 days
As CYP3A4 is considered to be one of the major CYP enzymes in intestinal metabolism, contributing to the clearance of various chemicals, we investigated whether provision of small-molecule compounds for 7 days also induced CYP3A4 (and CYP3A5) activity at the enzymatic level. To this end, HIOs were exposed for 24 h to the CYP3A4/5 substrate midazolam and subsequently the levels of 1′-OH-midazolam and 4-OH-midazolam were determined in HIO lysates and medium. 1′-OH-midazolam and 4-OH-midazolam levels were significantly higher after exposure to small-molecule compounds (Fig. 2f) indicating elevated CYP3A4/5 activity in these HIOs. Taken together, provision of small-molecule compounds for 7 days, after 28 days 3D culture, promotes differentiation of hiPSC-derived HIOs resulting in mature HIOs with CYP3A4/5 activity. Since this optimized protocol provides well-differentiated HIOs and more flexibility in culture, it was decided to continue the further studies using this optimized protocol (Fig. 2e).
Gene expression in HIOs, Caco-2 cells and the EpiIntestinal model
Gene expression of CYP genes and additionally some other genes coding for phase I biotransformation enzymes (carboxyl esterases (CEs)) and several common phase II biotransformation enzymes [uridine 5′-diphospho-glucuronosyltransferases (UGTs) and sulfotransferases (SULTs)] was compared between the HIOs cultured with the optimized protocol, the EpiIntestinal model, and Caco-2 cells. Comparison of the CYP genes showed significantly different expression of five out of nine genes between HIOs and the EpiIntestinal model, and of six out of nine genes between the Caco-2 cells and the EpiIntestinal model. The largest difference in CYP gene expression compared with the EpiIntestinal model was seen for CYP1B1 (530 times higher in HIOs), CYP1A2 (13 times higher in Caco-2 cells), CYP1A1 (7 times higher in Caco-2 cells), and CYP3A4 (7 times higher in HIOs) (Fig. 3a, Supplemental Table 5). When comparing the other genes for biotransformation enzymes, 7 out of 11 genes were differently expressed between HIOs and the EpiIntestinal model, and 9 out of 11 genes were differently expressed between Caco-2 cells and the EpiIntestinal model (Fig. 3b, Supplemental Table 5). Largest differences were observed for CES1, UGT1A8, UGT2B17 and UGT1A1 (125, 8, 6 and 4 times lower in HIOs, respectively) and for SULT1A1, SULT1A3, SULT1B1 and UGT1A10 (5, 3, 3 and 3 times higher in Caco-2 cells, respectively). Expression of the nuclear receptors aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR, NR1I3) and pregnane X receptor (PXR, NR1I2), known to regulate the expression of various CYPs (Tompkins and Wallace 2007; Zanger and Schwab 2013), was also compared between the different models. Gene expression of AHR and CAR was lower in HIOs as compared to the EpiIntestinal model and the Caco-2 cells, whereas mRNA levels of PXR were higher in HIOs than in the EpiIntestinal model and the Caco-2 cells (Fig. 3c, Supplemental Table 5). In summary, HIOs cultured with small-molecule compounds for 7 days, after 28 days 3D culture, showed similar or higher mRNA levels of CYP enzymes, and similar or lower mRNA levels of a selection of other biotransformation enzymes compared to the EpiIntestinal model (Fig. 3, Supplemental Table 5).
Gene expression in the EpiIntestinal model, HIOs and Caco-2 cells. a Relative expression of common intestinal CYP genes. b Relative expression of other selected biotransformation genes. c Relative expression of nuclear receptor genes. HIOs were exposed to small-molecule compounds for 7 days prior to mRNA extraction. Gene expression levels in the EpiIntestinal model were set at one. Asterisks indicate significant difference compared with the EpiIntestinal model (P < 0.05). Hashtags indicate significant difference between HIOs and Caco-2 cells (P < 0.05). Data are mean values ± SEM from triplicate wells
CYP induction in HIOs, Caco-2 cells and the EpiIntestinal model
To assess the inducibility of CYP enzymes by typical chemical CYP inducers in differentiated HIOs (HIOs exposed for 7 days to small-molecule compounds), Caco-2 cells and the EpiIntestinal model, all cell models were exposed to β-naphthoflavone, phenobarbital or rifampicin, which are typical inducers of CYP1A1/1A2/1B1, CYP2B6, and CYP3A4, respectively, in the human liver (Bernasconi et al. 2019). Relative gene expression levels compared to untreated controls (Supplemental Table 6) were determined and fold-changes were compared between the EpiIntestinal model, HIOs and Caco-2 cells (Fig. 4a). The typical CYP1 inducer β-naphthoflavone robustly induced expression of CYP1A1 and CYP1B1 in all models, but to the largest extent in Caco-2 cells (Fig. 4a). Interestingly, phenobarbital significantly induced CYP3A4 in the HIOs, but had no effect on CYP2B6 expression. No effect of phenobarbital was noticed in the Caco-2 cells or in the EpiIntestinal model (Fig. 4a). Rifampicin induced CYP3A4 in the HIOs and in the EpiIntestinal model (although the induction was not significant in the latter), but not in Caco-2 cells (Fig. 4a). None of the CYP inducers was found to induce expression of CYP2B6, CYP2C9, CYP2C19 or CYP2J2 in all three models (Fig. 4a).
CYP induction in the EpiIntestinal model, HIOs and Caco-2 cells. a Relative expression of common intestinal CYP genes in the EpiIntestinal model, Caco-2 cells and HIOs treated with 50 µM β-naphthoflavone, 500 µM phenobarbital or 50 µM rifampicin for 48 h. HIOs were exposed to small-molecule compounds for 7 days prior to exposure to CYP inducers. Gene expression levels in the control samples were set at one. Data are presented as mean ± SEM from triplicates. Asterisks indicate significantly different compared with CTRL. *P < 0.05, **P < 0.001. b Caco-2 cells and c HIOs treated with 50 µM rifampicin for 48 h and subsequently with 50 µM midazolam for 24 h. HIOs were exposed to small-molecule compounds for 7 days prior to exposure to rifampicin and the entire experiment was performed twice (three replicates per experiment). CYP3A4/5 activity was quantified by 1′-OH-midazolam and 4-OH-midazolam formation in cell lysates and medium. All single data points and the means (lines) are presented. Asterisks indicate significantly different compared with the CTRL. *P < 0.05
To assess whether the induction of CYP3A4 gene expression levels by rifampicin also translated in elevated CYP3A4 activity, HIOs (and Caco-2 cells) were exposed to rifampicin and subsequently to the CYP3A4/5 substrate midazolam. In Caco-2 cells, rifampicin did not induce formation of 1′-OH-midazolam and 4-OH-midazolam (Fig. 4b). Surprisingly, in HIOs 1′-OH-midazolam levels were also not significantly altered upon rifampicin pre-treatment, but a significant increase in 4-OH-midazolam formation was observed (Fig. 4c).
Discussion
In the present study, we improved an existing HIO differentiation protocol, leading towards better differentiation and expression of the most relevant intestinal CYP enzymes for xenobiotic metabolism without losing the capacity to keep them in prolonged culture, thus advancing throughput. Using this optimized protocol, we assessed the gene expression, induction and activity of important intestinal CYP enzymes in hiPSC-derived HIOs and compared these with the commonly used human colonic adenocarcinoma cell line Caco-2 and the human primary IEC-based EpiIntestinal model, closely resembling human small intestinal tissue (Ayehunie et al. 2018). The results of our study indicate that our HIO model expresses substantial levels of relevant intestinal CYP enzymes at the gene level, suggesting its relevance for in vitro biotransformation studies.
The results of the present study show that the gene expression of all evaluated CYPs, except CYP2B6 (and CYP2D6), increased upon treatment with the small-molecule compounds (A-83-01, PD98059, 5-aza-2′-deoxycytidine and DAPT), as reported before for CYP3A4 by Onozato et al. (2018). Therefore, to study intestinal metabolism and/or chemical toxicity, pre-treatment of HIOs with small compounds is recommended. These small compounds have all been reported to promote intestinal differentiation (Ogaki et al. 2013; Iwao et al. 2015; Macedo et al. 2018). Indeed, relative expression of intestinal differentiation markers, including markers for enteroendocrine and goblet cells, was elevated upon small-molecule compounds treatment. A drawback of addition of the small compounds, using the method as described by Onozato et al. (2018), is that the expansion of the HIO culture largely reduces, indicating that the increased differentiation is directly correlated with a reduced cell proliferation. This means that the HIOs cannot be kept in culture and a new batch of HIOs has to be started for each experiment. This largely hampers the throughput of the method, so we aimed to improve the existing protocol towards a protocol that can keep proliferating HIOs in culture, using the protocol as depicted in Fig. 1a, from which a subset of matured HIOs can be derived in a rather short period to perform an experiment, by treatment with small molecule compounds. To that end, small-molecule compounds were not added during differentiation from day 21 to 35, as reported by Onozato et al. (2018), but instead HIOs were allowed to differentiate for at least 35 days (28 days in 3D culture) and subsequently a subset was treated with small-molecule compounds for 3 or 7 days. Using the protocol with exposure to small compounds for 7 days, expression of CYP genes was similar as when using the original protocol of Onozato et al. (2018). Furthermore, also differentiation of the HIOs appeared to be improved compared with the protocol without addition of small compounds. This 7-day treatment period was therefore selected as optimal protocol for further experiments.
In HIOs obtained with our protocol, CYP3A4 gene expression was induced to a larger extent (25-fold) upon rifampicin treatment than reported for the HIO model by Onozato et al. (2018) (twofold). CYP3A4 was hardly expressed in our Caco-2 cells and was also not induced by rifampicin, which is in line with other studies (Martin et al. 2008; Ozawa et al. 2015; Negoro et al. 2016). Limited expression of the pregnane X receptor (PXR), responsible for CYP3A4 induction by rifampicin (Goodwin et al. 1999), has been described to be responsible for the lack of CYP3A4 induction by rifampicin in Caco-2 cells (Pfrunder et al. 2003; Sun et al. 2008). Indeed, PXR was limitedly expressed in Caco-2 cells, as evidenced by an average Ct value of 31, whereas PXR gene expression levels were tenfold higher in our HIO model. In addition, in our study CYP3A4 was also induced by phenobarbital in HIOs. Phenobarbital is a typical constitutive androstane receptor (CAR) agonist, resulting in CYP2B6 induction in the liver, which is used as a positive control in CYP induction studies in human hepatocytes (Bernasconi et al. 2019). The lack of CYP2B6 induction by phenobarbital in our HIO model suggests that a functional CAR may be lacking. The induction of CYP3A4 by phenobarbital has been shown to be mediated also by PXR in human liver HepaRG cells (Li et al. 2019). Therefore, the phenobarbital-mediated induction of CYP3A4 in HIOs, as measured in the present study, may be mediated by PXR as well.
To obtain insight whether CYP3A4 expression in HIOs is relevant regarding expression in the in vivo situation, HIOs were compared to the EpiIntestinal model, shown to closely resemble human small intestine tissue explants, also with regard to CYP3A4 mRNA expression (Ayehunie et al. 2018). Base level CYP3A4 expression in the EpiIntestinal model was approximately seven times lower than in the HIOs. Exposure of the EpiIntestinal model to either rifampicin or phenobarbital led to a non-significant elevation in CYP3A4 gene expression of approximately eight and two times, respectively. In the HIOs, a significant CYP3A4 induction amounting to approximately 25 and 15 times was observed upon rifampicin and phenobarbital exposure, respectively. Based on these analyses, CYP3A4 expression in the HIO model may be considered a bit high compared to the in vivo situation. However, the data should be interpreted with care due to complexity of comparing gene expression data from different models with also different expression levels of housekeeping genes. It must also be noted that interindividual differences in expression can be expected, and that the CYP3A4 expression in the EpiIntestinal model or in the HIO model used in the present study may be from a donor with relatively low or high expression, respectively.
Transcriptional activation of CYP1A and CYP1B is mediated by the AhR. The induction of CYP1A1 and CYP1B1 gene expression by β-naphthoflavone was elevated in all three models, but was highest in Caco-2 cells. Although gene expression of AHR was slightly lower in the HIOs as compared to Caco-2 cells and the EpiIntestinal model, CYP1B1 was, in comparison to the other two models, already highly expressed in HIOs prior to treatment with β-naphthoflavone, which may explain the relatively limited CYP1B1 induction upon β-naphthoflavone treatment in HIOs.
We next evaluated if the induction in CYP3A4 mRNA levels by rifampicin also translated into increased CYP3A4 activity. The conversion of midazolam into 1′-OH-midazolam is a commonly used method to determine CYP3A4/5 activity. In the literature, 1′-OH-midazolam hydroxylation of midazolam is often specifically linked to CYP3A4 activity, thereby neglecting CYP3A5 activity (Küblbeck et al. 2016; Yamaura et al. 2016; Jamwal et al. 2018; Onozato et al. 2018; Takayama et al. 2019). Albeit CYP3A4 is considered to be one of the most abundant intestinal CYP enzymes, the intestine also contains substantial CYP3A5 mRNA and protein levels (Canaparo et al. 2007; Peters et al. 2016). CYP3A4 and CYP3A5 mRNA levels were similar in our HIO model (exposed to small-molecule compounds for 7 days) (Supplemental Tables 4 and 5). Interestingly, although CYP3A4 expression was limited in our Caco-2 cells, CYP3A5 expression was substantially higher. To obtain more insight in relative CYP3A4 and CYP3A5 activities we determined the formation of both 1′-OH-midazolam and 4-OH-midazolam. Galetin et al. (2004) demonstrated, using CYP3A4 and CYP3A5 baculovirus-expressed recombinant systems, that the maximum velocity (Vmax) of 4-OH-midazolam formation was higher for CYP3A4 (2.5 pmol/min/pmol CYP3A4) than for CYP3A5 (0.5 pmol/min/pmol CYP3A5), and that the Vmax of 1′-OH-midazolam formation was higher for CYP3A5 (6.7 pmol/min/pmol CYP3A5) than for CYP3A4 (2.0 pmol/min/pmol CYP3A4), indicating a preference of CYP3A4 for 4-OH-midazolam formation and a preference of CYP3A5 for 1′-OH-midazolam formation. Exposing HIOs to rifampicin for 48 h did not alter 1′-OH-midazolam formation upon subsequent incubation with midazolam, but significantly increased 4-OH-midazolam formation. Together with the elevated gene expression levels of CYP3A4, but not of CYP3A5, these data suggest that rifampicin induces CYP3A4 activity in HIOs resulting in increased conversion of midazolam into 4-OH-midazolam. In Caco-2 cells, however, rifampicin did not increase 1′-OH-midazolam nor 4-OH-midazolam levels.
Although the expression of CYP3A4 mRNA and CYP3A4 activity are higher in HIOs than in Caco-2 cells, one cannot conclude that the HIO model outperforms the Caco-2 model in general. As shown in Fig. 3, gene expression of CYP1B1, CES1, SULT2A1 and UGT1A8 in Caco-2 cells resembled the gene expression in the EpiIntestinal model better than the expression in hiPSC-derived HIOs, suggesting that biotransformation of substrates of these enzymes may better be studied in Caco-2 cells than in the HIO model.
A striking difference in gene expression in the HIO model compared to the EpiIntestinal model was observed for CYP1B1, of which the expression was more than 500 times higher in the HIOs than in the EpiIntestinal model. CYP1B1 has been reported to play a role in the biotransformation of steroid hormones, fatty acids, vitamin A and melatonin (Li et al. 2017), and in the bioactivation of polycyclic aromatic hydrocarbons (Shimada et al. 1996). Although the present study does not indicate whether the high CYP1B1 mRNA expression also translates to a high CYP1B1 activity, one should consider this characteristic of the model. In general, this indicates that extensive characterization of any model is of utmost importance to understand its strong and weak characteristics and its related applicability domain.
Since our HIOs were obtained from hiPSCs from one donor, it would be of interest for future studies to perform experiments in hiPSCs-derived HIOs from different donors, to obtain insight in inter-individual differences in biotransformation enzyme expression, induction and activity. Furthermore, it would also be of interest to compare the hiPSC-derived HIO model with HIOs derived from adult stem cells like the model described by Pleguezuelos-Manzano et al. (2020), to obtain more insight into which model most closely represents the in vivo situation.
Taken together, in the current study we generated an optimized protocol for hiPSC-derived HIOs containing substantial gene expression of relevant intestinal CYP enzymes, showing that CYP1A1/1B1 gene expression can be induced by AhR activation and CYP3A4 gene expression by PXR activation, whereas CYP2B6 gene expression is not inducible, which may be due to a lack of functional CAR expression. Altogether, the data indicate that hiPSC-derived HIOs are useful models to study biotransformation of chemicals in the intestines.
Change history
25 February 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00204-021-03001-7
Abbreviations
- AhR:
-
Aryl hydrocarbon receptor
- CAR:
-
Constitutive androstane receptor
- CYP:
-
Cytochrome P450
- DE:
-
Definitive endoderm
- HG:
-
Hindgut endoderm
- IEC:
-
Intestinal epithelial cells
- hiPSCs:
-
Human induced pluripotent stem cells
- HIOs:
-
Human intestinal organoids
- PXR:
-
Pregnane X receptor
References
Aldhous MC, Shmakov AN, Bode J, Ghosh S (2001) Characterization of conditions for the primary culture of human small intestinal epithelial cells. Clin Exp Immunol 125:32–40
Ayehunie S, Landry T, Stevens Z, Armento A, Hayden P, Klausner M (2018) Human primary cell-based organotypic microtissues for modeling small intestinal drug absorption. Pharm Res 35:72
Bernasconi C, Pelkonen O, Andersson TB, Strickland J, Wilk-Zasadna I, Asturiol D, Cole T, Liska R, Worth A, Müller-Vieira U, Richert L, Chesne C, Coecke S (2019) Validation of in vitro methods for human cytochrome P450 enzyme induction: outcome of a multi-laboratory study. Toxicol In Vitro 60:212–228
Canaparo R, Finnström N, Serpe L, Nordmark A, Muntoni E, Eandi M, Rane A, Zara GP (2007) Expression of CYP3A isoforms and P-glycoprotein in human stomach, jejunum and ileum. Clin Exp Pharmacol Physiol 34:1138–1144
Cotreau MM, Von Moltke LL, Greenblatt DJ (2005) The influence of age and sex on the clearance of cytochrome P450 3A substrates. Clin Pharmacokinet 44:33–60
Eglen RM, Randle DH (2015) Drug discovery goes three-dimensional: goodbye to flat high-throughput screening? Assay Drug Dev Technol 13:262–265
Finkbeiner SR, Hill DR, Altheim CH, Dedhia PH, Taylor MJ, Tsai YH, Chin AM, Mahe MM, Watson CL, Freeman JJ, Nattiv R, Thomson M, Klein OD, Shroyer NF, Helmrath MA, Teitelbaum DH, Dempsey PJ, Spence JR (2015) Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Rep 4:1140–1155
Galetin A, Brown C, Hallifax D, Ito K, Houston JB (2004) Utility of recombinant enzyme kinetics in prediction of human clearance: impact of variability, CYP3A5, and CYP2C19 on CYP3A4 probe substrates. Drug Metab Dispos 32:1411–1420
Goodwin B, Hodgson E, Liddle C (1999) The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56:1329–1339
Grossmann J, Walther K, Artinger M, Kiessling S, Steinkamp M, Schmautz WK, Stadler F, Bataille F, Schultz M, Schölmerich J, Rogler G (2003) Progress on isolation and short-term ex-vivo culture of highly purified non-apoptotic human intestinal epithelial cells (IEC). Eur J Cell Biol 82:262–270
Hansten PD (2018) The underrated risks of tamoxifen drug interactions. Eur J Drug Metab Pharmacokinet 43:495–508
Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116:496–526
Iwao T, Kodama N, Kondo Y, Kabeya T, Nakamura K, Horikawa T, Niwa T, Kurose K, Matsunaga T (2015) Generation of enterocyte-like cells with pharmacokinetic functions from human induced pluripotent stem cells using small-molecule compounds. Drug Metab Dispos 43:603–610
Jamwal R, De La Monte SM, Ogasawara K, Adusumalli S, Barlock BB, Akhlaghi F (2018) Nonalcoholic fatty liver disease and diabetes are associated with decreased CYP3A4 protein expression and activity in human liver. Mol Pharm 15:2621–2632
Kato M, Chiba K, Hisaka A, Ishigami M, Kayama M, Mizuno N, Nagata Y, Takakuwa S, Tsukamoto Y, Ueda K, Kusuhara H, Ito K, Sugiyama Y (2003) The Intestinal first-pass metabolism of substrates of CYP3A4 and P-glycoprotein-quantitative analysis based on information from the literature. Drug Metab Pharmacokinet 18:365–372
Kosinski C, Stange DE, Xu C, Chan AS, Ho C, Yuen ST, Mifflin RC, Powell DW, Clevers H, Leung SY, Chen X (2010) Indian hedgehog regulates intestinal stem cell fate through epithelial–mesenchymal interactions during development. Gastroenterology 139:893–903
Küblbeck J, Hakkarainen JJ, Petsalo A, Vellonen KS, Tolonen A, Reponen P, Forsberg MM, Honkakoski P (2016) Genetically modified Caco-2 cells with improved cytochrome P450 metabolic capacity. J Pharm Sci 105:941–949
Le Guen L, Marchal S, Faure S, De Santa BP (2015) Mesenchymal–epithelial interactions during digestive tract development and epithelial stem cell regeneration. Cell Mol Life Sci 72:3883–3896
Li F, Zhu W, Gonzalez FJ (2017) Potential role of CYP1B1 in the development and treatment of metabolic diseases. Pharmacol Ther 178:18–30
Li L, Welch MA, Li Z, Mackowiak B, Heyward S, Swaan PW, Wang H (2019) Mechanistic insights of phenobarbital-mediated activation of human but not mouse pregnane X receptor. Mol Pharmacol 96:345–354
Lin JH, Lu AY (2001) Interindividual variability in inhibition and induction of cytochrome P450 enzymes. Annu Rev Pharmacol Toxicol 41:535–567
Lynch T, Price A (2007) The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am Fam Physician 76:391–396
Macedo MH, Araújo F, Martínez E, Barrias C, Sarmento B (2018) iPSC-derived enterocyte-like cells for drug absorption and metabolism studies. Trends Mol Med 24:696–708
Martin P, Riley R, Back DJ, Owen A (2008) Comparison of the induction profile for drug disposition proteins by typical nuclear receptor activators in human hepatic and intestinal cells. Br J Pharmacol 153:805–819
Mccracken KW, Howell JC, Wells JM, Spence JR (2011) Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc 6:1920–1928
Meran L, Baulies A, Li VSW (2017) Intestinal stem cell niche: the extracellular matrix and cellular components. Stem Cells Int 2017:7970385
Murray G, Barnes T, Sewell H, Ewen SW, Melvin WT, Burke MD (1988) The immunocytochemical localisation and distribution of cytochrome P-450 in normal human hepatic and extrahepatic tissues with a monoclonal antibody to human cytochrome P-450. Br J Clin Pharmacol 25:465–475
Negoro R, Takayama K, Nagamoto, Sakurai F, Tachibana M, Mizuguchi H (2016) Modeling of drug-mediated CYP3A4 induction by using human iPS cell-derived enterocyte-like cells. Biochem Biophys Res Commun 472:631–636
Ogaki S, Shiraki N, Kume K, Kume S (2013) Wnt and Notch signals guide embryonic stem cell differentiation into the intestinal lineages. Stem Cells 31:1086–1096
Onozato D, Yamashita M, Nakanishi A, Akagawa T, Kida Y, Ogawa I, Hashita T, Iwao T, Matsunaga T (2018) Generation of intestinal organoids suitable for pharmacokinetic studies from human induced pluripotent stem cells. Drug Metab Dispos 46:1572–1580
Ozawa T, Takayama K, Okamoto R, Negoro R, Sakurai F, Tachibana M, Kawabata K, Mizuguchi H (2015) Generation of enterocyte-like cells from human induced pluripotent stem cells for drug absorption and metabolism studies in human small intestine. Sci Rep 5:1–11
Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, Zeldin DC (2006) The human intestinal cytochrome P450 “Pie.” Drug Metab Dispos 34:880–886
Peters SA, Jones CR, Ungell AL, Hatley OJD (2016) Predicting drug extraction in the human gut wall: assessing contributions from drug metabolizing enzymes and transporter proteins using preclinical models. Clin Pharmacokinet 55:673–696
Pfrunder A, Gutmann H, Beglinger C, Drewe J (2003) Gene expression of CYP3A4, ABC-transporters (MDR1 and MRP1–MRP5) and hPXR in three different human colon carcinoma cell lines. J Pharm Pharmacol 55:59–66
Pleguezuelos-Manzano C, Puschhof J, van den Brink S, Geurts V, Beumer J, Clevers H (2020) Establishment and culture of human intestinal organoids derived from adult stem cells. Curr Protoc Immunol 130:1–24
Prueksaritanont T, Gorham L, Hochman J, Tran LO, Vyas KP (1996) Comparative studies of drug-metabolizing enzymes in dog, monkey, and human small intestines, and in Caco-2 cells. Drug Metab Dispos 24:634–642
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265
Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H (2011) Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141:1762–1772
Shimada T, Hayes CL, Yamazaki H, Amin S, Hecht SS, Guengerich FP, Sutter TR (1996) Activation of chemically diverse procarcinogens by human cytochrome P-450 1B1. Cancer Res 56:2979–2984
Spence JR, Mayhew CN, Rankin, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, Wells JM (2011) Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470:105–109
Stavropoulou E, Pircalabioru GG, Bezirtzoglou E (2018) The role of cytochromes P450 in infection. Front Immunol 9:1–7
Sun H, Chow ECY, Liu S, Du Y, Pang KS (2008) The Caco-2 cell monolayer: usefulness and limitations. Expert Opin Drug Metab Toxicol 4:395–411
Takayama K, Negoro R, Yamashita T, Kawai K, Ichikawa M, Mori T, Nakatsu N, Harada K, Ito S, Yamada H, Yamaura Y, Hirata K, Ishida S, Mizuguchi H (2019) Generation of human iPSC-derived intestinal epithelial cell monolayers by CDX2 transduction. Cell Mol Gastroenterol Hepatol 8:513–526
Tamminen K, Balboa D, Toivonen S, Pakarinen MP, Wiener Z, Alitalo K, Otonkoski T (2015) Intestinal commitment and maturation of human pluripotent stem cells is independent of exogenous FGF4 and R-spondin1. PLoS One 10:e0134551
Teo AKK, Ali Y, Wong KY, Chipperfield H, Sadasivam A, Poobalan Y, Tan EK, Wang ST, Abraham S, Tsuneyoshi N, Stanton LW, Dunn NR (2012) Activin and BMP4 synergistically promote formation of definitive endoderm in human embryonic stem cells. Stem Cells 30:631–642
Tompkins LM, Wallace AD (2007) Mechanisms of cytochrome P450 induction. J Biochem Mol Toxicol 21:176–181
Watson CL, Mahe MM, Múnera J, Howell JC, Sundaram N, Poling HM, Schweitzer JI, Vallance JE, Mayhew CN, Sun Y, Grabowski G, Finkbeiner SR, Spence JR, Shroyer NF, Wells JM, Helmrath MA (2014) An in vivo model of human small intestine using pluripotent stem cells. Nat Med 20:1310–1316
Xie F, Ding X, Zhang QY (2016) An update on the role of intestinal cytochrome P450 enzymes in drug disposition. Acta Pharm Sin B 6:374–383
Yamaura Y, Chapron BD, Wang Z, Himmelfarb J, Thummel KE (2016) Functional comparison of human colonic carcinoma cell lines and primary small intestinal epithelial cells for investigations of intestinal drug permeability and first-pass metabolism. Drug Metab Dispos 44:329–335
Zanger UM, Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141
Zomer P, Mol HGJ (2015) Simultaneous quantitative determination, identification and qualitative screening of pesticides in fruits and vegetables using LC-Q-OrbitrapTM-MS. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 32:1628–1636
Funding
This research was supported by the Dutch Ministry of Agriculture, Nature and Food Quality (Grant: KB37-001-003).
Author information
Authors and Affiliations
Contributions
Participated in research design: AWFJ, M van der Z, AACMP, JL. Conducted experiments: AWFJ, DR, LPMD. Performed data analysis: AWFJ, RN, LPMD, M van der Z, JL. Wrote or contributed to the writing of the manuscript: all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Janssen, A.W.F., Duivenvoorde, L.P.M., Rijkers, D. et al. Cytochrome P450 expression, induction and activity in human induced pluripotent stem cell-derived intestinal organoids and comparison with primary human intestinal epithelial cells and Caco-2 cells. Arch Toxicol 95, 907–922 (2021). https://doi.org/10.1007/s00204-020-02953-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-020-02953-6