Abstract
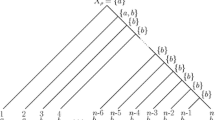
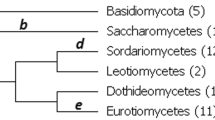
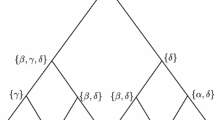
Information about protein sequences of ancestral organisms is important for identifying critical amino acid substitutions that have caused the functional change of proteins in evolution. Using computer simulation, we studied the accuracy of ancestral amino acids inferred by two currently available methods (maximum-parsimony [MP] and maximum-likelihood [ML] methods) in addition to a distance method, which was newly developed in this paper. All three methods give reliable inference when the divergence of amino acid sequences is low. When the extent of sequence divergence is high, however, the ML and distance methods give more accurate results than the MP method, particularly when the phylogenetic tree includes long branches. The accuracy of inferred ancestral amino acids does not change very much when a few present-day sequences are added or eliminated. When an incorrect model of amino acid substitution is used for the ML and distance methods, the accuracy decreases, but it is still higher than that for the MP method. When the tree topology used is partially incorrect, the accuracy in the correct part of the tree is virtually unaffected. The posterior probability of inferred ancestral amino acids computed by the ML and distance methods is an unbiased estimate of the true probability when a correct substitution model is used but may become an overestimate when a simpler model is used.
Similar content being viewed by others
References
Adachi J, Hasegawa M (1995) Improved dating of the human/ chimpanzee separation in the mitochondrial DNA tree: heterogeneity among amino acid sites. J Mol Evol 40:622–628
Cao Y, Aadachi J, Janke A, Pääbo S, Hasegawa M (1994) Phylogenetic relationships among eutherian orders estimated from inferred sequences of mitochondrial proteins: instability of a tree based on a single gene. J Mol Evol 39:519–527
Chandrasekharan UM, Sanker S, Glynias MJ, Karnik SS, Husain A (1996) Angiotensin II-forming activity in a reconstructed ancestral chymase. Science 271:502–505
Collins TM, Wimberger PH, Naylor GJP (1994) Compositional bias, character-state bias, and character-state reconstruction using parsimony. Syst Biol 43:482–496
Dayhoff MO, Schwartz RM, Orcutt BC (1978) A model of evolutionary change in proteins. In: Dayhoff MO (ed) Atlas of protein sequence and structure, vol 5, suppl 3. National Biomédical Research Foundation, Washington, DC, pp 345–352
Eck RV, Dayhoff MO (1966) Atlas of protein sequence and structure. National Biomedical Research Foundation, Silver Spring, MD
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1995) PHYLIP: phylogeny inference package. Version 3.57c. University of Washington, Seattle
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Hartigan JA (1973) Minimum evolution fits to a given tree. Biometrics 29:53–65
Jermann T, Opitz JG, Stackhouse J, Benner SA (1995) Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily. Nature 374:57–59
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Koshi JM, Goldstein RA (1996) Probabilistic reconstruction of ancestral protein sequences. J Mol Evol 42:313–320
KumarS, Tamura K, Nei M (1993) MEGA: molecular evolutionary genetics analysis, version 1.01. Pennsylvania State University, University Park
Lawson CL, Hanson RJ (1974) Solving least squares problems. Prentice-Hall, Englewood Cliffs, NJ, pp 158–165
Lee Y-H, Ota T, Vacquier VD (1995) Positive selection is a general phenomenon in the evolution of abalone sperm lysin. Mol Biol Evol 12:231–238
Libertini G, Di Donato A (1994) Reconstruction of ancestral sequences by the inferential method, a tool of protein engineering studies. J Mol Evol 39:219–229
Maddison WP (1995) Calculating the probability distributions of ancestral states reconstructed by parsimony on phylogenetic trees. Syst Biol 44:474–481
Maddison WP, Maddison DR (1992) MacClade: analysis of phylogeny and character evolution. Version 3. Sinauer, Sunderland, MA
Ota T, Nei M (1994) Estimation of the number of amino acid substitutions per site when the substitution rate varies among sites. J Mol Evol 38:642–643
Rzhetsky A, Nei M (1993) Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol Biol Evol 10: 1073–1095
Rzhetsky A, Nei M (1995) Tests of applicability of several substitution models for DNA sequence data. Mol Biol Evol 12:131–151
Saitou N, Nei M (1987) The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schluter D (1995) Uncertainty in ancient phylogenies. Nature 377:108–109
Yang Z (1994) Estimating the pattern of nucleotide substitution. J Mol Evol 39:105–111
Yang Z (1995) PAML: phylogenetic analysis by maximum likelihood. Version 1.1. Pennsylvania State University, University Park
Yang Z, Kumar S, Nei M (1995) A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141:1641–1650
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, J., Nei, M. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evol 44 (Suppl 1), S139–S146 (1997). https://doi.org/10.1007/PL00000067
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/PL00000067