Abstract
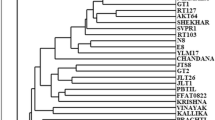
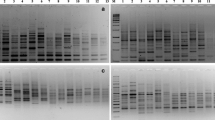
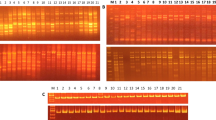
The determination of genetic differences among crop genotypes has become the primary need to grant patent and the protection of Plant Breeder Rights (PBR). In the present study RAPD and ISSR markers were employed for the characterization of 16 sesame genotypes. Twenty six RAPD and 17 ISSR primers that generated clear and reproducible banding patterns amplified 194 and 163 bands, respectively among 16 sesame genotypes. Both RAPD and ISSR primers showed maximum discrimination power, and produced putative variety specific bands, which could be used for the identification of all the sesame genotypes, individually. However, only AG and CA based ISSR primers were found effective in the discrimination of genotypes. A poor correlation was observed between the matrices produced by RAPD and ISSR primers, which might be due to the array of different sites of the genome. Though, there was greater similarity among sesame genotypes (0.78 for RAPD and 0.71 for ISSR), the observed genetic diversity (0.22 for RAPD and 0.29 for ISSR), was found effective for the characterization of sesame genotypes. It is suggested that putative variety specific RAPD and ISSR markers could be converted to Codominant sequence characterized amplified region/sequence tagged site (SCAR /STS) markers to develop robust variety specific markers.
Similar content being viewed by others
Abbreviations
- ISSR:
-
inter simple sequence repeat
- RAPID:
-
random amplified polymorphic DNA
- SCAR:
-
sequence characterized amplified region
- STS:
-
sequence tagged site
References
Johnson LA, Suleiman TM & Lusas EW, J Amer Oil Chem Soc, 56 (1979) 463.
Bedigian D, David S & Jack R, Biochem Sys Eco, 13 (1985) 133.
Bedigian D, Genet Resour Crop Evol, 50 (2003) 773.
Shiro I & Teruhisa U, Euphytica, 93 (1997) 375.
Kumar Vinod, Singh G, Sharma R & Sharma, SN, Indian J Plant Physiol, 12 (2007) 115.
Jana S & Pietrzak LN, Genetics, 119 (1988) 981.
Chengyin L, Weihua L & Mingjum L, J Tea Sci, 12 (1992) 15.
Jelinski D E & Cheliak WM, Am J Bot, 79 (1992) 728.
Botstein D, White RL, Skolnick MH & Davis RW, Am J Hum Genet, 32 (1980) 314.
Tanksley SD, Young ND, Paterson AH & Bonierbale MW, Biotechnology, 7 (1989) 257.
Miller JC & Tanksley SD, Theor Appl Genet, 80 (1990) 385.
Wang ZY, Second G & Tanksley SD, Theor Appl Genet, 83 (1992) 565.
Beckmann JS & Soller M, Euphytica, 35 (1986) 111.
Williams JGK, Kubelic AR, Livak KJ, Rafalski JA & Tingey SV, Nucleic Acids Res, 18 (1990) 6231.
Welsh J & McClelland M, Nucleic Acids Res, 18 (1990) 7213.
Zietkiewicz E, Rafalski A & Labuda D, Genomics, 20 (1994) 176.
Wu KS & Tanksley SD, Mol Gen Genet, 2411 (1993) 225.
Venkataramana Bhat K, Babraker PP & Lakhanpaul S, Euphytica, 110 (1999) 14.
Ercan GA, Taskin M & Turgut K, Genet Resour Crop Evol, 52 (2004) 599.
Kim M, Zur G, Danin-Poleg Y, Lee S, Shim K, Kang C & Kashi Y, Plant Breed, 121 (2002) 259.
Doyle JJ & Doyle JL, Focus, 12 (1990) 13.
Anderson JA, Churchill GA, Autroque JE, Tanksley SD & Swells ME, Genome, 36 (1993) 181.
Tessier C, David J, This P, Boursiquot JM & Charrier A. Theor Appl Genet, 98 (1999) 171.
Pavlicek A, Harda S & Flegr J, Folia Biol (Prague), 45 (1999) 97.
Awasthi AK, Nagaraja GM, Naik GV, Sriramana K, Thargavelu, K & Nagarju J, BMC Genetics, 141 (2004) 8.
Padmavathi N, Parminderjit K, AbuZafar B, George BT, Ray L & Umesh KR, In Proc XIII Intn Plant and Animal Genomes Conf, Town and Country Convention Centre, San Diego, CA (2005) p178.
Olufowote JO, Genome, 40 (1997) 370.
Sebastian LS, Hipolito LR & Garcia JS, J Breed Genet, 30 (1998) 73.
Ramakrishna W, Davierwala AP, Gupta VS & Ranjekar PK, Biochem Genet, 36 (1998) 323.
Morgante M & Olivieri, Am Plant J, 3 (1993) 175.
Lagercrantz U, Ellegren H & Andersson L, Nucleic Acids Res, 21 (1993) 1111.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharma, S.N., Kumar, V. & Mathur, S. Comparative Analysis of RAPD and ISSR Markers for Characterization of Sesame (Sesamum indicum L) Genotypes. J. Plant Biochem. Biotechnol. 18, 37–43 (2009). https://doi.org/10.1007/BF03263293
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF03263293