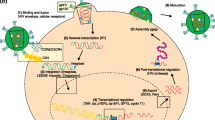
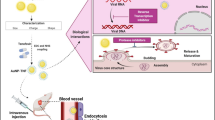
Abstract
Therapeutic strategies designed to treat HIV infection with combinations of antiviral drugs have proven to be the best approach for slowing the progression to AIDS. Despite the great success of highly active antiretroviral therapy (HAART), drug resistance and toxicity issues still remain a concern for some individuals. Therefore, alternative therapeutic strategies need to be developed to overcome these limitations. Nucleic acid-based therapeutics have been considered as an alternative to the currently used antivirals. In this regard, RNA interference (RNAi) can function as a gene-specific therapeutic option for controlling HIV-1 replication. Another type of therapeutic nucleic acid — aptamers — shows promise as a new and potent class of anti-HIV agent and can additionally function as a cell-type-specific delivery vehicle for targeted RNAi. The combined use of small interfering RNA (siRNAs) and aptamers could effectively block viral replication and prevent the emergence of resistant variants. In this review, we recapitulate recent progress and the therapeutic potential of aptamer-siRNA conjugates in the treatment of HIV infection.


Similar content being viewed by others
References
Richman DD, Margolis DM, Delaney M, et al. The challenge of finding a cure for HIV infection. Science 2009 Mar; 323(5919): 1304–7
Joshi PJ, Fisher TS, Prasad VR. Anti-HIV inhibitors based on nucleic acids: emergence of aptamers as potent antivirals. Curr Drug Targets Infect Disord 2003 Dec; 3(4): 383–400
Scherer L, Rossi JJ, Weinberg MS. Progress and prospects: RNA-based therapies for treatment of HIV infection. Gene Ther 2007 Jul; 14(14): 1057–64
Held DM, Kissel JD, Patterson JT, et al. HIV-1 inactivation by nucleic acid aptamers. Front Biosci 2006; 11: 89–112
Zhang Z, Blank M, Schluesener HJ. Nucleic acid aptamers in human viral disease. Arch Immunol Ther Exp 2004 Sep–Oct; 52(5): 307–15
Nielsen MH, Pedersen FS, Kjems J. Molecular strategies to inhibit HIV-1 replication. Retrovirology 2005; 2: 10
Chang HK, Gendelman R, Lisziewicz J, et al. Block of HIV-1 infection by a combination of antisense tat RNA and TAR decoys: a strategy for control of HIV-1. Gene Ther 1994 May; 1(3): 208–16
Banerjea A, Li MJ, Remling L, et al. Lentiviral transduction of Tar Decoy and CCR5 ribozyme into CD34+ progenitor cells and derivation of HIV-1 resistant T cells and macrophages. AIDS Res Ther 2004 Dec; 1(1): 2
Tuerk C, MacDougal S, Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A 1992 Aug; 89(15): 6988–92
Joshi P, Prasad VR. Potent inhibition of human immunodeficiency virus type 1 replication by template analog reverse transcriptase inhibitors derived by SELEX (systematic evolution of ligands by exponential enrichment). J Virol 2002 Jul; 76(13): 6545–57
Sayer N, Ibrahim J, Turner K, et al. Structural characterization of a 2′F-RNA aptamer that binds a HIV-1 SU glycoprotein, gp120. Biochem Biophys Res Commun 2002 May; 293(3): 924–31
Khati M, Schuman M, Ibrahim J, et al. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type1by gp120-binding 2′F-RNA aptamers. J Virol 2003 Dec; 77(23): 12692–8
Dey AK, Griffiths C, Lea SM, et al. Structural characterization of an antigp 120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA 2005 Jun; 11(6): 873–84
Cohen C, Forzan M, Sproat B, et al. An aptamer that neutralizes R5 strains of HIV-1 bindstocore residues of gp120 in the CCR5 binding site. Virology 2008 Nov; 381(1): 46–54
Zhou J, Swiderski P, Li H, et al. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res 2009 May; 37(9): 3094–109
Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998 Feb; 391(6669): 806–11
Zamore PD, Tuschl T, Sharp PA, et al. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000 Mar; 101(1): 25–33
Bennasser Y, Yeung ML, Jeang KT. RNAi therapy for HIV infection: principles and practicalities. Biodrugs 2007; 21(1): 17–22
Singh SK, Gaur RK. Progress towards therapeutic application of RNA interference for HIV infection. Biodrugs 2009; 23(5): 269–76
Tsygankov AY. Current developments in anti-HIV/AIDS gene therapy. Curr Opin Investig Drugs 2009 Feb; 10(2): 137–49
Berges BK, Rowan MR. The utilityofthe new generationofhumanized mice to study HIV-1 infection: transmission, prevention, pathogenesis, and treatment. Retrovirology 2011; 8: 65
Li MJ, Kim J, Li S, et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther 2005 Nov; 12(5): 900–9
Das AT, Brummelkamp TR, Westerhout EM, et al. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol 2004 Mar; 78(5): 2601–5
Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol 2007 Dec; 25(12): 1435–43
Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat Biotechnol 2007 Dec; 25(12): 1444–54
Boden D, Pusch O, Lee F, et al. Human immunodeficiency virus type 1 escape from RNA interference. J Virol 2003 Nov; 77(21): 11531–5
Schopman NC, ter Brake O, Berkhout B. Anticipating and blocking HIV-1 escape by second generation antiviral shRNAs. Retrovirology 2010; 7: 52
Zhou J, Li H, Li S, et al. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther 2008 Aug; 16(8): 1481–9
Neff CP, Zhou J, Remling L, et al. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med 2011 Jan; 3(66): 66ra6
Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001 May; 411(6836): 494–8
Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009 Jan; 457(7228): 426–33
Davidson BL, McCray Jr PB. Current prospects for RNA interference-based therapies. Nat Rev Genet 2011 May; 12(5): 329–40
Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov 2009 Feb; 8(2): 129–38
Goff SP. Knockdown screens to knockout HIV-1. Cell 2008 Oct; 135(3): 417–20
ter Brake O, Hooft K, Liu YP, et al. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther 2008 Mar; 16(3): 557–64
DiGiusto DL, Krishnan A, Li L, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med 2010 Jun; 2(36): 36ra43
Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science 2008 Feb; 319(5865): 921–6
Konig R, Zhou Y, Elleder D, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 2008 Oct; 135(1): 49–60
Zhou H, Xu M, Huang Q, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 2008 Nov; 4(5): 495–504
Yeung ML, Houzet L, Yedavalli VS, et al. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem 2009 Jul; 284(29): 19463–73
Juliano R, Alam MR, Dixit V, et al. Mechanisms and strategies for effective delivery ofantisense and siRNA oligonucleotides. Nucleic Acids Res 2008 Jul; 36(12): 4158–71
Perez-Martinez FC, Guerra J, Posadas I, et al. Barriers to non-viral vector-mediated gene delivery in the nervous system. Pharm Res 2011 Aug; 28(8): 1843–58
Wang J, Lu Z, Wientjes MG, et al. Delivery of siRNA therapeutics: barriers and carriers. AAPS J 2010 Dec; 12(4): 492–503
Mayer G. The chemical biology of aptamers. Angew Chem Int Ed Engl 2009; 48(15): 2672–89
Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov 2010 Jul; 9(7): 537–50
Syed MA, Pervaiz S. Advances in aptamers. Oligonucleotides 2010 Oct; 20(5): 215–24
Chaloin L, Lehmann MJ, Sczakiel G, et al. Endogenous expression of a high-affinity pseudoknot RNA aptamer suppresses replication of HIV-1. Nucleic Acids Res 2002 Sep; 30(18): 4001–8
Kraus E, James W, Barclay AN. Cutting edge: novel RNA ligands able to bind CD4 antigen and inhibit CD4+ T lymphocyte function. J Immunol 1998 Jun; 160(11): 5209–12
Zhou J, Rossi JJ. Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides 2011 Feb; 21(1): 1–10
Tan W, Wang H, Chen Y, et al. Molecular aptamers for drug delivery. Trends Biotechnol 2011; 29(12): 634–40
Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature 1990 Aug; 346(6287): 818–22
Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990 Mar; 344(6265): 467–8
Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990 Aug; 249(4968): 505–10
Kulbachinskiy AV. Methods for selection of aptamers to protein targets. Biochemistry 2007 Dec; 72(13): 1505–18
Guo KT, Paul A, Schichor C, et al. CELL-SELEX: novel perspectives of aptamer-based therapeutics. Int J Mol Sci 2008 Apr; 9(4): 668–78
Cerchia L, Duconge F, Pestourie C, et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS Biol 2005 Apr; 3(4): e123
Fang X, Tan W. Aptamers generated from Cell-SELEX for molecular medicine: a chemical biology approach. Acc Chem Res 2010 Jan; 43(1): 48–57
Zhou J, Rossi JJ. Aptamer-targeted cell-specific RNA interference. Silence 2010; 1(1): 4
Lupold SE, Hicke BJ, Lin Y, et al. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res 2002 Jul; 62(14): 4029–33
Dassie JP, Liu XY, Thomas GS, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol 2009 Sep; 27(9): 839–49
McNamara 2nd JO, Andrechek ER, Wang Y, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 2006 Aug; 24(8): 1005–15
Chu TC, Twu KY, Ellington AD, et al. Aptamer mediated siRNA delivery. Nucleic Acids Res 2006; 34(10): e73
Liu H, Rajasekaran AK, Moy P, et al. Constitutive and antibody-induced internalization of prostate-specific membrane antigen. Cancer Res 1998 Sep; 58(18): 4055–60
Hussey RE, Richardson NE, Kowalski M, et al. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature 1988 Jan; 331(6151): 78–81
Smith DH, Byrn RA, Marsters SA, et al. Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science 1987 Dec; 238(4834): 1704–7
Dalgleish AG, Beverley PC, Clapham PR, et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 1984 Dec-1985 Jan; 312(5996): 763–7
Kwong PD, Wyatt R, Robinson J, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998 Jun; 393(6686): 648–59
Sattentau QJ, Moore JP. The role of CD4 in HIV binding and entry. Philos Trans R Soc Lond B Biol Sci 1993 Oct; 342(1299): 59–66
Mukherjee S, Ghosh RN, Maxfield FR. Endocytosis. Physiol Rev 1997 Jul; 77(3): 759–803
Medina-Kauwe LK, Xie J, Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene Ther 2005 Dec; 12(24): 1734–51
Kim DH, Behlke MA, Rose SD, et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol 2005 Feb; 23(2): 222–6
Rose SD, Kim DH, Amarzguioui M, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res 2005; 33(13): 4140–56
Wheeler LA, Trifonova R, Vrbanac V, et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J Clin Invest 2011 Jun; 121(6): 2401–12
Zhou J, Neff CP, Swiderski P, et al. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with asticky bridge. Molecular Therapy. In press
Acknowledgements
This work is supported by grants from the National Institutes of Health AI29329, AI42552, and HL07470 awarded to J.J.R.
Author disclosures
The authors jointly own the US patent US 8,222,226 B2: ‘Cell-type specific aptamer-siRNA delivery system for HIV-1 therapy’. Issued date: July 17, 2012. The authors declare that they have no competing financial interests.
Author contributions
J.Z. drafted the article. J.J.R. revised it and gave final approval of the version to be published. All authors read and approved the final article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, J., Rossi, J.J. Therapeutic Potential of Aptamer-siRNA Conjugates for Treatment of HIV-1. BioDrugs 26, 393–400 (2012). https://doi.org/10.1007/BF03261896
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF03261896