Abstract
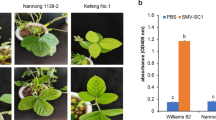
Soybean mosaic virus (SMV) is a severe disease in worldwide soybean production. A cross was made between Kefeng No. 1 with broad spectrum resistance to SMV and Nannong 1138–2, a susceptible cultivar. The inheritance of resistance to SMV strain Sa prevailing in southern China was analyzed. Results of x2 test from inoculation experiment on parents F1, F2 and F3 lines showed that the resistance to strain Sa was controlled by a single dominant gene Rsa. BSA method was adopted and 900 random 10-mer primers were used to amplify total DNA from resistant pool and susceptible pool in order to obtain polymorphic bands in two bulks. 16 primers could generate polymorphic bands, of which OPW-05 and OPAS-06 could generate the most stable RAPD patterns. RAPD markers OPW-05660 and OPAS-061800 were found to be linked to Rsa. Their order and genetic distance were OPAS-06180022.2cM Rsa10.1 cM OPW-05660. Southern blotting showed that both OPAS-061800 and 0PW-05660 were low copy DNA in genomic DNA. 0PW-05660 has been converted into an RFLP marker successfully. Additionally, pK644H, an RFLP marker, has been identified to be linked to 0PW-05660, and their genetic distance was 37.4 cM.
Similar content being viewed by others
References
Yu, P. G., Saghai, M. M. A., Buss, G. R.et al., RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance,Phytopathology, 1994, 84(1): 60.
Sambrook, J., Fritsch, E. F., Maniatis, T.,Molecular Cloning: A Laboratory Manual, 2nd ed., New York: Cold Spring Harbor Laboratory, 1989.
Michelmore, R. W., Paran, I., Kesseli, R. V., Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions using segregating populations,Proc. Natl. Acad. Sci., 1991, 88: 9828.
Verma, D. P. S., Shoemaker, R. C.,Soybean Genetics, Molecular Biology and Biotechnology, Wallingford: Cab International, 1995.
Kerm, P., Diers, B. W., Olson, T. C.et al., RFLP mapping in soybean: association between marker loci and variation in quantitative traits,Genetics, 1990, 126: 735.
Diers, B. W., Fehr, W., Keim, P.et al., RFLP analysis of soybean seed protein and oil content,Theor. Appl. Genet., 1992, 83: 608.
Skorupska, H. T., Shoemaker, R. C., Warner, A.et al., Restriction fragment length polymorphism in soybean germplasm of the Southern USA,Crop Sci., 1993, 33: 1169.
Lark, K. G., Weisemann, J. M., Matthews, B. F.et al., A genetic map of soybean (Glycine max L.) using an intraspecific cross of two cultivars: “Minsoy and Noir 1”,Theor. Appl. Genet., 1993, 86: 901.
Shoemaker, R. C., Specht, J. E., Integration of the soybean molecular and classical genetic linkage groups,Crop Sci., 1995, 35: 436.
Author information
Authors and Affiliations
About this article
Cite this article
Zhang, Z., Chen, S. & Gai, J. Molecular markers linked to Rsa resistant to soybean mosaic virus. Chin. Sci. Bull. 44, 154–158 (1999). https://doi.org/10.1007/BF02884741
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF02884741