Summary
It seems as though nature was most innovative at the very beginning of life on this Earth a few billion years ago. For example, the functional competence of most, if not all, of the sugar-metabolizing enzymes was clearly established before the division of eukaryotes from prokaryotes eons ago, each critical active-site amino acid sequence being conserved ever since by bacteria as well as by mammals. I contend that this initial innovativeness was due to the first set of coding sequences being repeats of base oligomers, thus encoding polypeptide chains of various periodicities; such periodical polypeptide chains can easily acquire α-helical and β-sheet-forming segments. In fact, the entire length of sugarmetabolizing enzymes is comprised of alternating α-helical and β-sheet-forming segments.
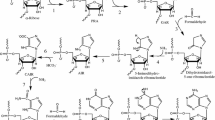
In the prebiotic (therefore nonenzymatic) replication of nucleic acids, what was in short supply was long templates, for there apparently was no inherent obstacle in copying of long templates, if such existed, in the presence of Zn2+. I submit that in this prebiotic condition, only those nucleotide oligomers that were internal doubles were automatically assured of progressive elongation to become long templates. For example, a decamer that was a pentameric repeat and its complementary sequence may pair unequally to initiate the next round of replication: first unit pairing with second, and a paired segment serving as a primer. As a consequence of this unequal pairing, decameric templates managed to become pentadecameric templates only after one round of replication, and this elongation process had no inherent limit.
Similar content being viewed by others
References
Bridson PK, Orgel LE (1980) Catalysis of accurate poly (C) disrected synthesis of 3–5′ linked oligoguanytes by Zn+2. J Mol Biol 144: 567–577
Dayhoff MO (ed) (1972) Atlas of protein sequences and structure. National Biomedical Research Foundation. Silver Spring MD
Dyson F (1985) Origins of life. Cambridge University Press, Cambridge, UK
Eigen M, Schuster P (1977) The hypercycle: a principle of natural self-organization. Part A: emergence of hypercycle. Naturwissenschaften 64: 541–565
Hill D, Blumenthal T (1983) Does Qβ replicase, synthesize RNA in the absence of templates? Nature 301: 350–352
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University press, Cambridge, UK
Kioshita S, Negoro S, Murayama M, Bisaria VS, Sawada S, Okada H (1977) 6-Amino hexanoic acid cyclic dimer hydrolase. A new cyclic amide hydrolase produced byAcromobacter guttatus KI 72. Eur J Biochem 80: 489–495
Kinoshita S, Terada T, Taniguchi T, Takene Y, Masuda S, Okada H (1981) Purification and characterization of 6-aminohexonic-acid-oligomer hydrolase ofFlavobacterium sp. KI72. Eur J Biochem 116: 547–551
McKean D, Huppi K, Bell M, Staudt L, Gerhard W, Weigert M (1984) Generation of antibody diversity in the immune response of BALB/C mice to influenza virus hemagglutinins. Proc Natl Acad Sci USA 811: 3180–8184
Mezquita J, Connor W, Winkfein RJ, Dixon GH (1985) An H1 histone gene from rainbow trout (Salmo gairdnerii). J Mol Evol 21: 209–219
Miller SL, Orgel LE (1974) The origin of life on the Earth. Prentice-Hall, New York
Ohno S (1984) The birth of a unique new enzyme from an alternate reading frame of the preexisted, internally repetitious coding sequences. Proc Natl Acad Sci USA 81: 2421–2425
Okada H, Negoro S, Kimura H, Nakamura S (1983) Evolutionary adaptation of plasmid-encoded enzymes for degrading nylon oligomers. Nature 306: 203–206
Ycas M (1974) On earlier states of biochemical system. J Theor Biol 44: 145–160
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ohno, S. Evolution from primordial oligomeric repeats to modern coding sequences. J Mol Evol 25, 325–329 (1987). https://doi.org/10.1007/BF02603117
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02603117