Abstract
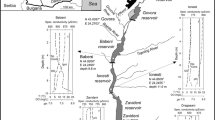
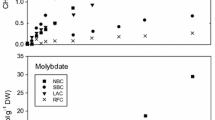
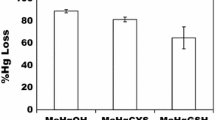
The role of biological activities in the reduction and volatilization of Hg(II) from a polluted pond was investigated. Elemental mercury was evolved from pond water immediately following spiking with203Hg(NO3)2, whereas an acclimation period of 36 hours was required in control samples collected from a nearby, unpolluted river before onset of volatilization. Genes encoding the bacterial mercuric reductase enzyme (mer genes) were abundant in DNA fractions extracted from biomass of the pond microbial community, but not in samples extracted from control communities. Thus, evolution of Hg0 was probably due to activities mediated by the bacterial mercuric reductase. Of four characterizedmer operons, the system encoded by transposon 501 (mer(Tn501)) dominated and likely contributed to the majority of the observed Hg(II) volatilization. Thus,mer-mediated reduction and volatilization could be used to reduce Hg(II) concentrations in polluted waters, in turn decreasing rates of methylmercury formation by limiting substrate availability.
Similar content being viewed by others
References
American Public Health Association (1989) Standard methods for the examination of water and wastewater. 17th Edition. Washington DC
Barkay T (1987) Adaptation of aquatic microbial communities to Hg2+ stress. Appl Environ Microbiol 53:2725–2732.
Barkay T, Gillman M, Liebert C (1990) Genes encoding mercuric reductases from selected gram-negative aquatic bacteria have a low degree of homology withmerA of transposon 501. Appl Environ Microbiol 56:1695–1701.
Barkay T, Liebert C Gillman M (1989) Hybridization of DNA probes with whole-community genome for detection of genes that encode microbial responses to pollutants:mer genes and Hg2+ resistance. Appl Environ Microbiol 55:1574–1577.
Begley T, Walts A, Walsh C (1986) Mechanistic studies of a protonolytic organomercurial cleaving enzyme: Bacterial organomercurial lyase. Biochemistry 25:7192–7200
Ben-Bassat D, Mayer AM (1978) Light-induced Hg volatilization and O2 evolution inChlorella and the effect of DCMU and methylamine, Physiol Plant 42:33–38
Berman M, Chase T Jr, Bartha R (1990) Carbon flow in mercury biomethylation byDesulfovibrio desulfuricans. Appl Environ Microbiol 56:298–300
Björnberg A, Håkanson L, Lundbergh K (1988) A theory on the mechanisms regulating the bioavailability of mercury in natural waters. Environ Pollut 49:53–61.
Bloom N (1989) Determination of picogram levels of methylmercury by aqueous phase ethylation, followed by cryogenic gas chromatography with cold vapour atomic fluorescence detection. Can J Fish Aquat Sci 46:1131–1140
Bloom NS, Fitzgerald WF (1988) Determination of volatile mercury species at the picogram level by low temperature gas chromatography with cold-vapor atomic fluorescence detection. Anal Chim Acta 209:151–161
Braman RS, Johnson DL (1974) Selective absorption tubes and emission technique for determination of ambient forms of mercury in air, Environ Sci Technol 8:996–1003
Hobbie JE, Daley RJ, Jasper S (1977) Use of Nucleopore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol 33:1225–1228
Jain RK, Burlage RS, Sayler GS (1988) Methods for detecting recombinant DNA in the environment. Crit Rev Biotechnol 8:33–84
Jernelöv A, Lann H (1973) Studies in Sweden on feasibility of some methods for restoration of mercury-contaminated bodies of water. Environ Sci Technol 7:712–718
Kusano T, Guangyong J, Inoue C Silver S (1990) Constitutive synthesis of a transport function encoded by theThiobacillus ferrooxidans merC gene cloned inEscherichia coli. J Bacteriol 172:2688–2692
Landner L (1971) Biochemical model for the biological methylation of mercury suggested from methylation studies in vivo withNeurospora crassa. Nature (London) 230:452–454
Liebert C, Barkay, T, Turner RR (1991) Acclimation of aquatic microbial communities to Hg(II) and CH3HgCl in polluted freshwater ponds. Microb Ecol 21:139–149
Nakamura K, Sakamoto M, Uchiyama H, Yagi O (1990) Organomercurial-volatilizing bacteria in the mercury-polluted sediment of Minamata Bay, Japan, Appl Environ Microbiol 56:304–305
Nelson JD, Colwell RR (1975) The ecology of mecury-resistant bacteria in Chesapeake Bay. Microb Ecol 1:191–218
Ogram A, Sayler GS, Barkay T (1987) The extraction and purification of microbial DNA from sediments. J Microbiol Methods 7:57–66
Olson BH, Lester JN, Cayless SM, Ford S (1989) Distribution of mercury resistance determinants in bacterial communities of river sediments. Water Res 23:1209–1217
Pennacchioni A, Marchetti R, Gaggino GF (1976) Inability of fish to methylate mercuric chloride in vivo. J Environ Qual 5:451–454
Rudd JWM, Turner MA (1983) The English-Wabigoon river system: II. Suppression of mercury and selenium bioaccumulation by suspended and bottom sediments. Can J Fish Aquat Sci 40:2218–2227
Rudd JWM, Turner MA, Furutani A, Swick AL, Townsend BE (1983) The English-Wabigoon river system: I. A synthesis of recent research with a view towards mercury amelioration. Can J Fish Aquat Sci 40:2206–2217
Schroeder WH, Jackson R (1984) An instrumental analytical technique for vapour-phase mercury species in air, Chemosphere 13:1041–1051
Silver S, Misra TK (1988) Plasmid-mediated heavy metal resistances. Ann Rev Microbiol 42: 717–743
Steffan RJ, Korthals ET, Winfrey MR (1988) Effects of acidification on mercury methylation, demethylation, and volatilization in sediments from an acid-susceptible lake. Appl Environ Microbiol 54:2003–2009
Summers AO, Barkay T (1989) Metal resistance genes in the environment. In: Levy SB, Miller RV (eds) Gene transfer in the environment. McGraw-Hill Publishing Co., New York, pp 287–308
Summers AO, Sugarman LI (1974) Cell-free mercury(II)-reducing activity in a plasmid-bearing strain ofEscherichia coli. J Bacteriol 119:242–249
Thomas JM, Ward CH (1989) In situ biorestoration of organic contaminants in the subsurface. Environ Sci Technol 23:760–766
Turner MA, Rudd JWM (1983) The English-Wabigoon river system: III. Selenium in lake enclosures: Its geochemistry, bioaccumulation, and ability to reduce mercury bioaccumulation. Can J Fish Aquat Sci 40:2228–2240
Turner RR, VandenBrook AJ, Barkay T, Elwood JW (1989) Volatilization, methylation, and demethylation of mercury in a mercury-contaminated stream. In: Vernet JP (ed) Proc Intl Conf on Heavy Metals in the Environment, CEP Consultants Ltd, Edinburgh UK, pp 353–356
Weiss AA, Murphy SD, Silver S (1977) Mercury and organomercurial resistances determined by plasmids inStaphylococcus aureus. J Bacteriol 132:197–208
Winfrey MR, Rudd JWM (1990) Environmental factors affecting the formation of methylmercury in low pH lakes, Environ Toxicol Chem 9:853–869
Xun L, Campbell NER, Rudd JWM (1987) Measurements of specific rates of net methyl mercury production in the water column and surface sediments of acidified and circumneutral lakes. Can J Fish Aquat Sci 44:750–757
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Barkay, T., Turner, R.R., VandenBrook, A. et al. The relationships of Hg(II) volatilization from a freshwater pond to the abundance ofmer genes in the gene pool of the indigenous microbial community. Microb Ecol 21, 151–161 (1991). https://doi.org/10.1007/BF02539150
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02539150