Abstract
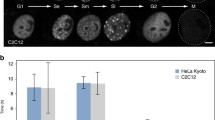
Newly synthesized DNA in mammalian nuclei is concentrated in discrete nuclear granules called replication foci. These foci may be visualized using antibodies against 5-bromodeoxyuridine. In the early S-phase cells 100–250 foci are usually detected. On average, individual foci range between 0.5 and 2 µm in diameter and can be seen as clusters of more than ten average-sized (60–100 kb) synchronously activated replicons. In this study, employing minor modifications of the previous methods, we report the visualization of small replication foci of about 0.3 µm diameter (mini-foci). Some foci are clustered into folded chains consisting of 2–40 subunits. DNA content of one mini-focus is estimated to be 50–120 kb and there are 500–1500 mini-foci per cell in the early S-phase. Experimentally induced decrease in replicon size does not affect the size of mini-foci, suggesting that these represent elementary units of DNA replication in mammalian nuclei and are probably identical to the basic structural DNA loop domains.
Similar content being viewed by others
References
Cleaver J (1978) DNA repair and its coupling to DNA replication in eukaryotic cells.Biochim Biophys Acta 516: 489–493.
Cook PR, Brasell IA, Jost E (1976) Characterization of nuclear structures containing superhelical DNA.J Cell Sci 22: 303–324.
Edenberg HJ, Huberman JA (1975) Eukaryotic chromosome replication.Annu Rev Genet 9: 245–284.
Hancock R, Boulikas T (1982) Functional organization in the nucleus.Int Rev Cytol 79: 165–214.
Hand R (1978) Eukaryotic DNA: organization of the genome for replication.Cell 15: 317–325.
Hassan AB, Cook PR (1993) Visualization of replication sites in unfixed human cells.J Cell Sci 105: 541–550.
Hozak P, Hassan AB, Jackson DA, Cook PR (1993) Visualization of replication factories attached to a nucleoskeleton.Cell 73: 361–373.
Huberman JA, Riggs AD (1968) On the mechanism of DNA replication in mammalian chromosomes.J Mol Biol 32: 327–341.
Jackson DA, Dickinson P, Cook PR (1990) the size of chromatin loops in HeLa cells.EMBO J 9: 567–571.
Kill IR, Bridger JM, Campbell KHS, Maldonado-Codina G, Hutchinson CJ (1991) The timing of the formation and usage of replicase clusters in S-phase nuclei of human diploid fibroblasts.J Cell Sci 100: 869–876.
Nakamura H, Morita T, Sato C (1986) Structural organization of replicon domains in eukaryotic cell nucleus.Exp Cell Res 165: 291–297.
Nakayasu H, Berezney R (1989) Mapping replicational sites in the eukaryotic cell nucleus.J Cell Biol 108: 1–11.
O'Keefe RT, Henderson SC, Spector DL (1992) Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences.J Cell Biol 116: 1095–1110.
Svetlova M, Solovjeva L, Stein G, Chamberland C, Vig B, Tomilin N (1994) The structure of human S-phase chromosome fibres.Chromosome Res 2: 47–52.
Taylor JH (1977) Increase in DNA replication sites in cells held at the beginning of S-phase.Chromosoma 57: 341–350.
Taylor JH, Hozier JC (1976) Evidence for a four micron replication unit in CHO cells.Chromosoma 57: 341–350.
Tomilin N, Rosanov Y, Zenin V, Bozhkov V, Vig B (1993) A new and rapid method for visualising DNA replication in spread DNA by immunofluorescence detection of incorporated 5-iododeoxyuridine.Biochem Biophys Res Commun 190: 257–262.
Trask BJ, Pinkel D, van der Engh GJ (1989) The proximity of DNA sequences in interphase cell nuclei is correlated to genomic distance and permits ordering of cosmids spanning 250 kilobase pairs.Genomics 5: 710–717.
Trask BJ, Massa HF, Kenwrick S, Gitschier J (1991) Mapping of human chromosome Xq28 by two-color fluorescencein situ hybridization of DNA sequences to interphase cell nuclei.Am J Hum Genet 48: 1–15.
Tschurikov NA, Ponomarenko NA (1992) Detection of DNA domains inDrosophila, human and plant chromosomes possessing mainly 50 to 150 kilobase stretches of DNA.Proc Natl Acad Sci USA 89: 6751–6755.
van der Engh GJ, Sachs R, Trask BJ (1992) Estimating genomic distance from DNA sequence location in cell nuclei by a random walk model.Science 257: 1410–1412.
van Dierendonk JH, Keyzer R, Velde CJH, Corneliss CH (1989) Subdivision of S-phase by analysis of nuclear 5-bromodeoxyuridine staining patterns.Cytometry 10: 143–150.
Vogelstein B, Pardoll DM, Coey DS (1980) Supercoiled loops and eukaryotic DNA replication.Cell 22: 79–85.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Tomilin, N., Solovjeva, L., Krutilina, R. et al. Visualization of elementary DNA replication units in human nuclei corresponding in size to DNA loop domains. Chromosome Res 3, 32–40 (1995). https://doi.org/10.1007/BF00711159
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00711159