Abstract
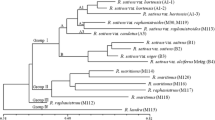
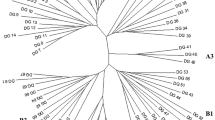
One-hundred-and-eighty-one nuclear DNA probes were used to examine restriction-fragment length polymorphism in inbred lines of the cultivated sunflower (Helianthus annuus L.). The probes were from six libraries: two genomic libraries — one made with PstI and the other with HindIII, and four cDNA libraries — from etiolated plantlets, green leaves, ovaries, petals and anthers. Total DNA from 17 inbred lines representing an overview of the genetic stocks of sunflower, including restorer and maintainer lines of the classical cytoplasmic male sterility, was digested with four different restriction enzymes and probed in 331 probe-enzyme combinations. Of 181 clones analysed, 73 probes were found to be polymorphic. Genetic distances between inbreds were calculated from the resultant proportion of shared bands and submitted to principal component analysis and the UPGMA ‘tree-making’ method. The RFLP analysis allowed a clear differentiation between restorer and maintainer lines of the cytoplasmic male sterility, together with a grouping of some of the genotypes from the same origin. The analysis of the accuracy of distance estimation as a function of the number of probe-enzyme combinations used, indicates that 40–50 combinations ensure a confidence level of near 95%. Considering the inbreds as representatives of the range of cultivated inbreds, estimates of gene diversity, as well as estimates of average gene diversity between and within the sets of restorer and maintainer lines, were calculated. Estimation of gene diversity showed that the available genetic variability in cultivated sunflower, based on allelic frequencies, is lower than that of other plants (H=0.20). Moreover, we show that the proportion of genetic variability due to the difference between maintainer and restorer lines (Dm) is about 2%.
Similar content being viewed by others
References
Berry ST, Allen RJ, Barnes SR, Caligari PDS (1994) Restriction fragment length polymorphism between inbred lines of cultivated sunflower (Helianthus annuus L.). Theor Appl Genet 89:435–441
Choumane W, Heizmann P (1988) Structure and variability of nuclear ribosomal genes in the genus Helianthus. Theor Appl Genet 76:481–489
Fatokun CA, Danesh D, Young ND, Stewart EL (1993) Molecular taxonomic relationships in the genus Vigna based on RFLP analysis. Theor Appl Genet 86:97–104
Gentzbittel L, Nicolas P (1990) Improvement of ‘a BASIC program to construct evolutionary trees from restriction endonuclease data’ with the use of Pascal. J Hered 81:491–492
Gentzbittel L, Perrault A, Nicolas P (1992) Molecular phylogeny of the Helianthus genus, based on nuclear restriction-fragment length polymorphism (RFLP). Mol Biol Evol 9:872–892
Goffreda JC, Burnquist WB, Beer SC, Tanksley SD, Sorrells ME (1992) Application of molecular markers to assess genetic relationships among accessions of wild oat, Avena sterilis. Theor Appl Genet 85:146–151
Havey MJ, Muehlbauer FJ (1989) Variability for restriction-fragment lengths and phytogenies in lentil. Theor Appl Genet 77:839–843
Helentjaris T, King G, Slocum M, Siedenstrang C, Wegman S (1985) Restriction-fragment length polymorphisms as probes for plant diversity and their development as tools for applied plant breeding. Plant Mol Biol 5:109–118
Kiss T, Solymosy F (1987) Isolation of high-molecular-weight plant nuclear DNA suitable for use in recombinant DNA technology. Acta Biochim Biophys Hung 22:1–5
Kochert G, Halward T, Branch WD, Simpson CE (1991) RFLP variability in peanut (Arachis hypogaea L.) cultivars and wild species. Theor Appl Genet 81:565–570
Korell G, Mosges F, Friedt W (1992) Construction of a sunflower pedigree map. Helia 15–17:7–16
Leclercq P (1969) Une stérilité mâle cytoplasmique chez le Tournesol. Ann Amélior Plant 19:99–106
McGrath JM, Quiros CF (1992) Genetic diversity at isozyme and RFLP loci in Brassica campestris as related to crop type and geographical origin. Theor Appl Genet 83:783–790
Melchinger AE, Boppenmaier J, Dhillon BS, Pollmer WG, Herrmann RG (1992). Genetic diversity for RFLPs in European maize in breds. II. Relation to performance of hybrids within, versus between, heterotic groups for forage traits. Theor Appl Genet 84:672–681
Miller JC, Tanksley SD (1990). RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor Appl Genet 80:437–448
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Li W-H (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nei M, Roychoudhury AK (1982) Genetic relationship and evolution of human races. Evol Biol 14:1–59
Nodari RO, Koinange EMK, Kelly JD, Gepts P (1992) Towards an integrated linkage map of common bean. 1. Development of genomic DNA probes and levels of restriction-fragment length polymorphism. Theor Appl Genet 84:186–192
Quillet MC, Vear F, Branlard G (1992) The use of isozyme polymorphism for identification of sunflower (Helianthus annuus) inbred lines. J Genet Breed 46:295–304
Rieseberg LH, Soltis DE, Palmer JD (1988) A molecular reexamination of introgression between Helianthus annuus and H bolanderi (Compositae). Evolution 42:227–238
Rieseberg LH, Beckstrom-Sternberg S, Doan K (1990) Helianthus annuus ssp. texanus has chloroplast DNA and nuclear ribosomal RNA genes of Helianthus debilis ssp cucumerifolius. Proc Natl Acad Sci USA 87:593–597
Schilling EE, Heiser CB (1981) Infrageneric classification of Helianthus (compositae). Taxon 30:393–403
Serieys H, Vincourt P (1987) Caractérisation de nouvelles sources de stérilité mâle chez le Tournesol. Les colloques de l'INRA no 45, INRA, Paris, pp 53–64
Van de Ven WTG, Duncan N, Ramsay G, Phillips M, Powell W, Waugh R (1993) Taxonomic relationships between V.faba and its relatives based on nuclear and mitochondrial RFLPs and PCR analysis. Theor Appl Genet 86:71–80
Author information
Authors and Affiliations
Additional information
Communicated by J. W. Snape
Rights and permissions
About this article
Cite this article
Gentzbittel, L., Zhang, YX., Vear, F. et al. RFLP studies of genetic relationships among inbred lines of the cultivated sunflower, Helianthus annuus L.: evidence for distinct restorer and maintainer germplasm pools. Theoret. Appl. Genetics 89, 419–425 (1994). https://doi.org/10.1007/BF00225376
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00225376