Abstract
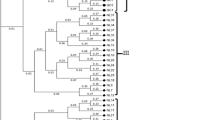
Genetic relationships were studied by means of ten isoenzymatic systems, at the genus and species level, using two distances and four methods of aggregation in a germplasm collection of 198 cultivars and accessions of 54 species belonging to Citrus and 13 related genera. The most consistent results were obtained by the chord distance and the neighbor-joining clustering method. Citrus species were distributed in two main groups: the orange-mandarin group and the lime lemon-citron-pummelo group. The species C. halimii and C. tachibana are not included in these groups. Mandarin species fall into three main subgroups: one includes C. sinensis; the second, C. aurantium, the third, small-fruit species. The citron, the pummelo and the ancient lemon subgroups form a cluster to which the species belonging to subgenus Papeda and the cultivated limes, lemons and bergamots are related. Microcitrus spp, to which Severinia buxifolia and Atalantia ceylanica seem to be related, cluster with the lime lemon-citron-pummelo group while Fortunella is close to the orange-mandarin group. Poncirus trifoliata, the most important species for citrus rootstock improvement is located far from Citrus but connected to it through Fortunella spp. A broad distribution of species has been found that should be taken into account to sample new genotypes in the search of desired characters in order to fully and efficiently use genetic resources for citrus improvement.
Similar content being viewed by others
References
Asíns MJ, Herrero R, Navarro L (1995) Factors affecting Cirtus tree isozyme-gene expression. Theor Appl Genet 90:892–898
Barrett HC, Rhodes AM (1976) A numerical taxonomic study of affinity relationships in cultivated Cirus and its close relatives. Syst Bot 1:105–136
Benzecri JP (1970) Distance distributionelle et metrique du chi-deux en analyse factorielle des correspondences. Laboratoire de Statistique Mahtematique, Paris
Blondel L (1978) Classification botanique des espèces du genre Citrus. Fruit 33:695–720
Bretó MP, Asíns MJ, Carbonell EA (1993) Genetic variability in Lycopersicon species and their genetic relationships. Theor Appl Genet 86:113–120
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Evolution 21:550–570
Esen A, Scora RW (1977) Amylase polymorphism in Citrus and some related genera. Am J Bot 64:305–309
Frankel OH (1990) Germplasm conservation and utilization in horticulture. In: Bennett AB, O'Neill SD (eds) Horticulture biotechnology. Wiley-Liss, New York, pp 5–17
Green RM, Vardi A, Galun E (1986) The plastome of Citrus. Physical map, variation among Citrus cultivars and species and comparison with related genera. Theor Appl Genet 72:170–177
Grosser JW, Gmitter FG Jr (1990) Wide-hybridization of Citrus via protoplast fusion: progress, strategies and limitations. In: Bennett AB, O'Neill SD (eds) Horticultural biotechnology. Wiley-Liss, New York, pp 31–41
Handa T, Oogaki C (1985) Numerical taxonomic study of Citrus L. and Fortunella Swingle using morphological characters. J Jpn Soc Hortic Sci 54:145–154
Handa T, Ishizawa Y, Oogaki C (1986) Phylogenetic study of fraction I progtectein study of fraction I protein in the genus Citrus and its close related genera. Jpn J Gen 61:15–24
He SW, Liu GF, Li WP (1988) Wild mandarin oranges in China. In: Goren R, Mendel K (eds) Proc Int Soc Citricult, vol 1. Balaband, Rehovot, Israel and Margrat Publishers, Weikersheim, Germany, pp 113–121
Herrero R, Asíns MJ, Carbonell EA, Navarro L (1996) Genetic diversity in the orange subfamily Aurantioideae. I. Intraspecies and intragenus genetic variability. Theor Appl Genet 92:599–609
Hirai M, Kozaki I, Kajiura I (1986) Isozyme analysis and phylogenetic relationships of citrus. Jpn J Breed 36:377–389
Hirai M, Mitsue S, Kita K, Kajiura I (1990) A survey and isozyme analysis of wild mandarin, tachibana (Citrus tachibana (Mak.) Tanaka) growing in Japan. J Jpn Soc Hortic Sci 59:1–7
Hodgson RW (1967) Horticultural varieties of citrus. In: Reuther W, Webber HJ, Batchelor LD (eds) The citrus industry, vol 1. University of California, Berkeley, pp 431–591
Iwamasa M, Nito N (1988) Cytogenetics and the evolution of modern cultivated citrus. In: Goren R, Mendel K (eds) Proc Int Soc Citricult, vol 1. Balaband, Rehovot, Israel and Margrat Publishers, Weikersheim, Germany, pp 265–275
Iwamasa M, Nito N, Ling J-T (1988) Intra- and intergeneric hybridization in the orange subfamily, Aurantioideae. In: Goren R, Mendel K (eds) Proc Int Soc Citricult, vol 1. Balaband, Rehovot, Israel and Margrat Publishers, Weikersheim, Germany, pp 123–130
Kozaki I, Hirai M (1981) Pollen ultrastructure of citrus cultivars. In: Matsumoto K (eds) Proc Int Soc Citricult, vol 1: I.S.C. Printing Co., Misaki-cho, Japan, pp 19–22
Lapointe F-J, Legendre P (1992) Statistical significnance of the matrix correlation coefficient for comparing independent phylogenetic trees. Syst Biol 41:378–384
Luro F, Laigret F, Bové JM, Ollitrault P (1992) Application of random amplified polymorphic DNA (RAPD) to citrus genetics and taxonomy. In: Tribulato E, Gentile A, Reforgiato (eds) Proc Int Soc Citricult, vol 1. I.S.C., MCS Congress. Catania, Italy, pp 225–228
Malik MN, Scora RW, Soost RK (1974) Studies on the origin of the lemon. Hilgardia 42:361–382
Navarro L, Juarez J, Pina JA, Ballester LF, Arregui JM (1988) The citrus variety improvement program in Spain after eleven years. In: Goren R, Mendel K (eds) Proc 10th Conf Int Organization Citrus Virol IOCV. Balaband, Rehovot, Israel and Margrat Publishers, Weikersheim, Germany, pp 400–406
Nei M (1991) Relative efficiencies of different tree-making methods for molecular data. In: Miyamoto MM, Cracraft J (eds) Phylogenetic analysis of DNA sequences. Oxford University Press, Oxford, pp 90–128
Ollitrault P (1990) Isozymes and restriction fragment length polymorphisms as genetic markers in citrus selection. In: Proc 4th Asia-Pacific Int Conf Citrus Rehabilitation. FAO-UNDP RAS /86/022 regional project. FAO, Rone, pp 59–63
Potvin C, Bergeron Y, Simon JP (1983) A numerical taxonomic study of selected citrus species (Rutaceae) based on biochemical characters. Syst Bot 8:127–133
Prim RC (1957) Shortest connection matrix network and some generalisations. Bell System Technol J 36:1389–1401
Rahman MM, Nito N, Isshiki S (1994) Genetic analysis of phosphoglucoisomerase isozymes in “true citrus fruit trees”. Sci Hortic 60:17–22
Roose ML (1988) Isozymes and DNA restriction fragment polymorphism in citrus breeding and systematics. In: Goren R, Mendel K (eds) Proc Int Soc Citricult, vol 1. Balaband, Rehovot, Israel and Margrat Publishers, Weikersheim, Germany, pp 155–165
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Scora RW (1988) Biochemistry, taxonomy and evolution of modern cultivated citrus. In: Goren R, Mendel K (eds) Proc Int Soc Citricult, vol 1. Balaband, Rehovot, Israel and Margrat Publishers, Weikersheim, Germany, pp 277–289
Scora W, Kumamoto J (1983) Chemotaxonomy of the genus Citrus. In: Waterman PG, Grundon MF (eds) Chemistry and chemical taxonomy of the rutales. (Phytochem Soc Eur Symp Ser No 22.) Academic Press, London, pp 342–351
Sneath PHA, Sokal RR (1973) Numerical taxonomy. Freeman. San Francisco
Stone BC, Lowry JB, Scora RW, Jong K (1974) Citrus halimii: A new species from Malaya and Peninsular Thailand. Biotropica 5:102–110
Swingle WT, Reece PC (1967) The botany of citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD (eds) The citrus industry, vol 1. University of California, Berkeley, pp 190–430
Swofford DL, Olsen GJ (1990) Phylogeny reconstruction. In: Hillis DM, Moritz C (eds) Molecular systematics. Sinauer Assoc, Sunderland, Mass., pp 411–501
Torres AM, Soost RK, Diedenhofen U (1978) Leaf isoenzymes as genetic markers in citrus. Am J Bot 65:869–881
Ward JH (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244
Yamamoto M, Kobayashi S, Nakamura Y, Yamada Y (1993) Phylogenetic relationships of citrus revealed by RFLP analysis of mitochondrial and chloroplast DNA. Jpn J Breed 43:355–365
Ye YM, Kong Y, Zheng XH (1981) Studies on pollen morphology of citrus plants. In: Matsumoto K (ed) Proc Int Soc Citricult, vol 1. I.S.C. Printing Co., Misaki-cho, Japan, pp 23–25
Author information
Authors and Affiliations
Additional information
Communicated by P. M. A. Tigerstedt
Rights and permissions
About this article
Cite this article
Herrero, R., Asíns, M.J., Pina, J.A. et al. Genetic diversity in the orange subfamily Aurantioideae. II. Genetic relationships among genera and species. Theoret. Appl. Genetics 93, 1327–1334 (1996). https://doi.org/10.1007/BF00223466
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00223466