Abstract
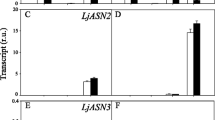
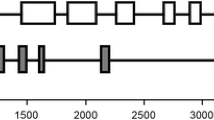
Two cDNA clones, LJAS1 and LJAS2, encoding different asparagine synthetases (AS) have been identified and sequenced and their expression in Lotus japonicus characterised. Analysis of predicted amino acid sequences indicated a high level of identity with other plant AS sequences. No other AS genes were detected in the L. japonicus genome. LJAS1 gene expression was found to be root-enhanced and lower levels of transcript were also identified in photosynthetic tissues. In contrast, LJAS2 gene expression was root-specific. These patterns of AS gene expression are different from those seen in pea. AS gene expression was monitored throughout a 16 h light/8 h dark day, under nitrate-sufficient conditions. Neither transcript showed the dark-enhanced accumulation patterns previously reported for other plant AS genes. To evaluate AS activity, the molecular dynamics of asparagine synthesis were examined in vivo using 15N-ammonium labelling. A constant rate of asparagine synthesis in the roots was observed. Asparagine was the most predominant amino-component of the xylem sap and became labelled at a slightly slower rate than the asparagine in the roots, indicating that most root asparagine was located in a cytoplasmic ‘transport’ pool rather than in a vacuolar ‘storage’ pool. The steady-state mRNA levels and the 15N-labelling data suggest that light regulation of AS gene expression is not a factor controlling N-assimilation in L. japonicus roots during stable growth in N-sufficient conditions.
Similar content being viewed by others
References
Andrews M: The partitioning of nitrate assimilation between root and shoot of higher plants. Plant Cell Environ 9: 511–519 (1986).
Chattopadhyay N, Kher R, Godbole M: Inexpensive SDS/phenol method for RNA extraction from tissues. BioTechniques 15: 24–26 (1993).
Church GM, Gilbert W: Genomic sequencing. Proc Natl Acad Sci USA 81: 1991 (1984).
Davies KM, King GA: Isolation and characterization of a cDNA clone for a harvest-induced asparagine synthetase from Asparagus officinalis L. Plant Physiol 102: 1337–1340 (1993).
Fortin MG, Purohit SK, Verma DPS: The primary structure of soybean (Glycine max) ubiquitin is identical to other plant ubiquitins. Nucl Acids Res 16: 11377 (1988).
Grundemann D, Koepsaell H: Ethidium bromide staining during glyoxalation with glyoxal for sensitive detection of RNA in agarose gel electrophoresis. Anal Biochem 216: 459–461 (1994).
Handeberg K, Stougaard J: Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2: 487–496, (1992).
Huber TA, Streeter JG: Purification and properties of asparagine synthetase from soybean root nodules. Plant Sci 42: 9–17 (1985).
Jang J-C, Sheen J: Sugar-sensing in higher plants. Plant Cell 6: 1665–1679 (1994).
Joy KW, Ireland RJ, Lea PJ: Asparagine synthetase in pea leaves, and the occurrence of an asparagine synthetase inhibitor. Plant Physiol 73: 165–168 (1983).
Lam H-M, Peng SS-Y, Coruzzi GM: Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol 106: 1347–1357 (1994).
Lea PJ, Miffin BJ: Transport and metabolism of asparagine and other nitrogen compounds within the plant. In: Stumpf PK, Conn EE (eds) The Biochemistry of Plants, pp. 569–607. Academic Press, New York (1980).
Lee RB: The release of nitrite from barley roots in response to metabolic inhibitors, uncoupling agents, and anoxia. J Exp Bot 30: 119–133 (1979).
Lee RB, Lewis MJ: Synthesis and 15N-labelling of glutamine and glutamate in maize roots during early stages of 15N-ammonium assimilation. J Exp Bot 45: 767–777 (1994).
Li X-Z, Larson DE, Glibetic M, Oaks A: Effect of glutamine on the induction of nitrate reductase. Physiol Plant 93: 740–744 (1995).
Mei B, Zalkin H: A cysteine-histidine-aspartate catalytic triad is involved in glutamine amide transfer function in purF-type glutamine amidotransferases. J Biol Chem 264: 16613–16619 (1989).
Murray MJ, Thompson WF: Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8: 4321–4325 (1980).
Oaks A: A re-evaluation of nitrogen assimilation in roots. BioScience 42: 103–111 (1992).
Rothnie HM, Reid J, Hohn T: The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′-end formation in plants. EMBO J 13: 2200–2210 (1994).
Rouze P, Caboche M: Nitrate reduction in higher plants: molecular approaches to function and regulation. In: Wray JL (ed) Inducible Plant Proteins, pp. 45–77. Society for Experimental Biology Seminar Series. Cambridge University Press, Cambridge, UK (1992).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1989).
Scott DB, Farnden KJF, Robertson JG: Ammonia assimilation in lupin nodules. Nature 263: 703–705 (1976).
Sheen J: Metabolic repression of transcription in higher plants. Plant Cell 2: 1027–1038 (1990).
Sieciechwicz KA, Joy KW, Ireland RJ: The metabolism of asparagine in plants. Phytochemistry 27: 663–671 (1988).
Streeter JM: Asparaginase and asparagine transaminase in soybean leaves and nodules. Plant Physiol 69: 848–852 (1977).
Thomas BR, Rodriguez RL: Metabolite signals regulate gene expression and source/sink relations in cereal seedlings. Plant Physiol 106: 1235–1239 (1994).
Tsai F-Y, Coruzzi GM: Dark-induced and organ-specific expession of two asparagine synthetase genes in Pisum sativum. EMBO J 9: 323–332 (1990).
Tsai F-Y, Coruzzi GM: Light repression transcription of asparagine synthetase genes in photosynthetic and non-photosynthetic organs of plants. Mol Cell Biol 11: 4966–4972 (1991).
Urquhart AA, Joy KW: Transport, metabolism, and redistribution of xylem-borne amino acids in developing pea shoots. Plant Physiol 69: 1226–1232 (1982).
Verwoerd TC, Dekker BMM, Hoekema A: A small-scale procedure for the rapid isolation of plant RNAs. Nucl Acids Res 17: 2362 (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Waterhouse, R.N., Smyth, A.J., Massonneau, A. et al. Molecular cloning and characterisation of asparagine synthetase from Lotus japonicus: Dynamics of asparagine synthesis in N-sufficient conditions. Plant Mol Biol 30, 883–897 (1996). https://doi.org/10.1007/BF00020801
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00020801