Abstract
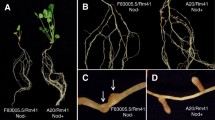
Specificity in legume-Rhizobium symbiosis depends on plant and rhizobial genes. As our objective was to study broad host-range determinants of rhizobia, we sought a legume and a Rhizobium with the lowest possible specificity. By inoculating 12 different legumes with a heterogenous collection of 35 fast-growing rhizobia, we found Rhizobium sp. NGR234 to be the Rhizobium and Vigna unguiculata to be the plant with the lowest specificities. Transfer of cloned fragments of the Sym-plasmid pNGR234a into heterologous rhizobia, screening for extension of host-range of the transconjugants to include V. unguiculata, and restriction mapping of the Hsn- and overlapping clones, proved that there were at least three distinct Hsn-regions (HsnI, II, and III) on pNGR234a. HsnI is located next to nodD, HsnII is linked to nifKDH and HsnIII to nodC. In addition to nodulation of Vigna, HsnI conferred upon the transconjugants the ability to nodulate Glycine max, Macroptilium atropurpureum and Psophocarpus tetragonolobus. All three Hsn-regions, when transferred to the appropriate recipients, induced root-hair-curling on M. atropurpureum. Hsn-region III was able to complement a mutation in the host-range gene nodH of R. meliloti strain 2011. Homology to “nod-box”-sequences could be shown only for the sub-clones containing HsnII and HsnIII, thus suggesting different regulation mechanisms for HsnI and HsnII/III.
Similar content being viewed by others
References
Bachem CWB, Banfalvi Z, Kondorosi E, Schell J, Kondorosi A: Identification of host range determinants in the Rhizobium species MPIK3030. Mol Gen Genet 203: 42–48 (1986).
Bachem CWB, Kondorosi E, Banfalvi Z, Horvath B, Kondorosi A, Schell J: Identification and cloning of nodulation genes from the wide host range Rhizobium strain MPIK3030. Mol Gen Genet 199: 271–278 (1985).
Bagdasarian MM, Amann E, Lurz R, Rückert B, Bagdasarian M: Activity of the hybrid trp-lac (tac) promoter of E. coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene 26: 273–282 (1983).
Bassam BJ, Rolfe BG, Djordjevic MA: Macroptilium a-tropurpureum (siratro) host specificity genes are linked to a nodD-like gene in the broad host range Rhizobium strain NGR234. Mol Gen Genet 203: 49–57 (1986).
Broughton WJ, Bohlool BB, Shaw CH, Bohnert HJ, Pankhurst CE: Conserved plasmid/chromosome sequences in fast- and slow-growing rhizobia that nodulate the same plant. Arch Microbiol 141: 14–21 (1985).
Broughton WJ, Dilworth MJ: Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 (1971).
Broughton WJ, Heycke N, Meyer z.A. H, Pankhurst CE: Plasmid-linked nif and “nod” genes in fast-growing rhizobia that nodulate Glycine max, Psophocarpus tetragonolobus, and Vigna unguiculata. Proc Natl Acad Sci USA 81: 3093–3097 (1984).
Broughton WJ, John CK: Rhizobia in tropical legumes. III. Experimentation and supply in Malaysia — 1927 to 1976. In: Broughton WJ, John CK, Rajarao JC, Lim B (eds) Soil Microbiology and Plant Nutrition, pp 113–136. Kuala Lumpur: University of Malaya Press (1979).
Broughton WJ, Wong CH, Lewin A, Samrey U, Myint H, Meyer z.A. H, Dowling DN, Simon R: Identification of Rhizobium plasmid sequences involved in recognition of Psophocarpus, Vigna, and other legumes. J Cell Biol 102: 1173–1182 (1986).
Buikema WJ, Long SR, Brown SM, van denBos RC, Earl DC, Ausubel FM: Cosmid cloning of a large region of symbiotic genes from the megaplasmid of Rhizobium meliloti. J Mol Appl Genet 2: 249–260 (1983).
Chua KJ, Pankhurst CE, MacDonald PE, Hopcroft DH, Jarvis BDW, Scott DB: Isolation and characterisation of transposon Tn5-induced symbiotic mutants of Rhizobium loti. J Bacteriol 162: 335–343 (1985).
Debellé F, Rosenberg C, Vasse J, Maillet F, Martinez E, Dénarié J, Truchet J: Assignment of symbiotic developmental phenotypes to common and specific nodulation (nod) genetic loci of Rhizobium meliloti. J Bacteriol 168: 1075–1086 (1986).
Debellé F, Sharma SB: Nucleotide sequence of Rhizobium meliloti RCR2011 genes involved in host specificity of nodulation. Nucleic Acids Res 14: 7453–7472 (1986).
Denhardt DT: A membrane filter technique for the detection of complementary DNA. Biochem Biophys Res Comm 23: 641–646 (1966).
Djordjevic MA, Innes RW, Wijffelman CA, Schofield PR, Rolfe BG: Nodulation of specific legumes is controlled by several distinct loci in Rhizobium trifolii. Plant Mole Biol 6: 389–401 (1986).
Djordjevic MA, Schofield PR, Ridge RW, Morrison NA, Bassam BJ, Plazinski J, Rolfe BG: Rhizobium nodulation genes involved in root hair curling (Hac) are functionally conserved. Plant Mol Biol 4: 147–160 (1985).
Downie JA, Hombrecher G, Ma Q-S, Knight CD, Wells B, Johnston AWB: Cloned nodulation genes of Rhizobium leguminosarum determine host-range specificity. Mol Gen Genet 190: 359–365 (1983).
Egelhoff TT, Fisher RF, Jacobs TW, Mulligan JT, Long SR: Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodA, B, C. DNA 4: 241–248 (1985).
Fuhrmann M, Hennecke H: Nitrogenase Fe protein gene (nifH). J Bacteriol 158: 1005–1011 (1984).
Glauert AM, Kerridge D, Horne RW: The fine structure and mode of attachment of the sheathed flagellum of Vibrio metchnikovii. J Cell Biol 18: 327–336 (1963).
Haas D, Holloway BW: Chromosome mobilisation by the R plasmid R68.45. A tool in Pseudomonas genetics. Mol Gen Genet 158: 229–237 (1978).
Horvath B, Kondorosi E, John M, Schmidt J, Török J, Györgypac Z, Barabas J, Wieneke U, Schell J, Kondorosi A: Organization, structure and symbiotic functions of Rhizobium meliloti nodulation genes determining host specificity for Alfalfa. Cell 46: 335–343 (1986).
Innes RW, Kuempel PL, Plazinski J, Canter-Cremers H, Rolfe BG, Djordjevic MA: Plant factors induce expression of nodulation and host range genes in Rhizobium trifolii. Mol Gen Genet 281: 426–432 (1985).
Ish-Horowicz D, Burke JF: Rapid and efficient cosmid cloning. Nucleic Acids Res 9: 2989–2998 (1981).
Jacobs TW, Egelhoff TT, Long SR: Physical and genetic map of a Rhizobium meliloti nodulation gene region and nucleotide sequence of nodC. J Bacteriol 162: 469–476 (1985).
Jordan DC, Allen ON: Rhizobiaceae. In: Buchanan RE, Gibbons NE (eds) Bergey's Manual of Determinative Bacteriology, 8th edn, pp 261–264. Baltimore: The Williams & Williams Co. (1974).
Kaluza K, Hennecke H: Fine structure analysis of the nifDK operon encoding the α and β subunits of dinitrogenase from Rhizobium japonicum. Mol Gen Genet 196: 35–42 (1984).
Kondorosi E, Banfalvi Z, Kondorosi A: Physical and genetic analysis of a symbiotic region of Rhizobium meliloti: identification of nodulation genes. Mol Gen Genet 193: 445–452 (1984).
Kondorosi E, Kondorosi A: Nodule induction on plant roots by Rhizobium. Trends in Biol Sci 11: 295–299 (1986).
Lamb JW, Regensburger B, Fischer H-M, Göttfert M, Meyer L, Ebling S, Studer H, Hahn M, Hennecke H: Bradyrhizobium japonicum genes, involved in soybean root nodule development. In: Lugtenberg B (ed) Recognition in Microbe-Plant Symbiotic and Pathogenic Interaction, pp 79–86. Heidelberg: Springer (1986).
Lim G: Rhizobium and nodulation of legumes in Singapore. In: Broughton WJ, John CK, Rajarao JC, Lim B (eds) Soil Microbiology and Plant Nutrition, pp 159–175. Kuala Lumpur: University of Malaya Press (1979).
Long SR, Buikema WJ, Ausubel FM: Cloning of R. meliloti nodulation genes by direct complementation of Nod--mutants. Nature 298: 485–488 (1982).
Maniatis T, Fritsch EF, Sambrook J: Molecular Cloning — A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press (1982).
't Mannetje L. A re-examination of the taxonomy of the genus Rhizobium and related genera using numerical analysis. Antonie van Leeuwenhoek 33: 477–491 (1967).
Meynell E, Cooke M: Repressor minus and operator constitutive derepressed mutants of F-like R factors. Their effect on chromosomal transfer by HfrC. Genet Res 14: 309–313 (1969).
Mulligan JT, Long SR: Induction of Rhizobium meliloti nodC expression by plant exudate requires nodD. Proc Natl Acad Sci USA 82: 6609–6613 (1985).
Nelson LM, Child JJ: Nitrogen fixation and hydrogen metabolism by Rhizobium leguminosarum isolates in pea root nodules. Canad J Microbiol 27: 1028–1034 (1981).
Norris DO: A red strain of Rhizobium from Lotononis bainesii Baker. J Agric Res 9: 629–632 (1958).
Norris DO: Rhizobium relationships in legumes. In: proceedings of the Ninth International Grassland Congress, pp 1087–1092 (1965).
Pankhurst CE: Symbiotic effectiveness of antibiotic-resistant mutants of fast- and slow-growing strains of Rhizobium nodulating Lotus species. Can J Microbiol 23: 1026–1033 (1977).
Pankhurst CE, Broughton WJ, Bachem C, Kondorosi E, Kondorosi A: Identification of nitrogen fixation and nodulation genes on a large plasmid from a broad host-range Rhizobium sp. In: Pühler A (ed) Molecular Genetics of the Bacteria-Plant Interaction, pp 169–176. Heidelberg/Berlin: Springer-Verlag (1983).
Rosenberg C, Boistard P, Dénarié J, Casse-Delbart F: Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol Gen Genet 184: 326–333 (1981).
Rossen L, Johnston AWB, Downie JA: DNA sequence of the Rhizobium leguminosarum nodulation genes nodAB and C required for root hair curling. Nucleic Acids Res 12: 9497–9508 (1985).
Rossen L, Shearman CA, Johnston AWB, Downie JA: The nodD gene of Rhizobium leguminosarum is autoregulatory and in the presence of plant exudate induces the nodA, B, C genes. EMBO J 4: 3369–3373 (1985).
Rostas K, Kondorosi E, Horvath B, Simoncsits A, Kondorosi A: Conservation of extended promotor regions of nodulation genes in Rhizobium. Proc Natl Acad Sci USA 83: 1757–1761 (1986).
Schofield PR, Ridge RW, Rolfe BG, Shine J, Watson JM: Host-specific nodulation is encoded on a 14 kb DNA fragment in Rhizobium trifolii. Plant Mol Biol 3: 3–11 (1984).
Schofield PR, Watson JM: DNA sequence of Rhizobium trifolii nodulation gene reveals a reiterated and potentially regulatory sequence preceding nodABC and nodFE. Nucleic Acids Res 14: 2891–2903 (1986).
Shearman CA, Rossen L, Johnston AWB, Downie JA: The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl-carrier protein and is regulated by nodD plus a factor in pea root exudate. EMBO J 5: 647–652 (1986).
Sundaresan V, Jones JDG, Ow DW, Ausubel FM: Klebsiella pneumonia nifA product activates the Rhizobium meliloti nitrogenase promotor. Nature 301: 728–731 (1983).
Török J, Kondorosi E, Stepkowski T, Pósoai J, Kondorosi A: Nucleotide sequence of Rhizobium meliloti nodulation genes. Nucleic Acids Res 12: 9509–9524 (1985).
Trinick MJ: Nodulation of tropical legumes. I. Specificity in the Rhizobium symbiosis of Leucaena leucocephala. Exp Agric 4: 243–253 (1968).
Trinick MJ: Relationships amongst the fast-growing Rhizobium of Lablab purpureus, Leucaena leucocephala, Mimosa sp., Acacia farnesiana, and Sesbania grandiflora and their affinities with other Rhizobium groups. J Appl Bacteriol 49: 39–53 (1980).
Trinick MJ, Galbraith J: The Rhizobium requirements of the non-legume Parasponia in relationship to the cross-inoculation group concept of legumes. New Phytol 86: 17–26 (1980).
Williams WM, Broughton WJ: Measurement of acetylene reduction in various systems. In: Broughton WJ, John CK, Rajarao JC, Lim B (eds) Soil Microbiology and Plant Nutrition, pp 289–339. Kuala Lumpur: University of Malaya Press (1979).
Wong CH, Pankhurst CE, Kondorosi A, Broughton WJ: Morphology of root nodules and nodule-like structures formed by Rhizobium and Agrobacterium strains containing a Rhizobium meliloti megaplasmid. J Cell Biol 97: 787–794 (1983).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Lewin, A., Rosenberg, C., H. Meyer, z.A. et al. Multiple host-specificity loci of the broad host-range Rhizobium sp. NGR234 selected using the widely compatible legume Vigna unguiculata . Plant Mol Biol 8, 447–459 (1987). https://doi.org/10.1007/BF00017990
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00017990