Abstract
Dragonflies are colorful diurnal insects with large compound eyes. Because they visually recognize conspecific and heterospecific individuals, their body color plays essential roles in ecology and reproductive biology. Here I introduce the recent topics of molecular mechanisms underlying color vision and color formation in dragonflies. Complex wing color polymorphism is recognized among the two closely related Japanese Mnais species, presumably due to stepwise character displacement to avoid interspecific mating. We discovered an extraordinary large number of visual opsin genes by RNA sequencing of 12 dragonfly species. Manual correction after de novo assembly was crucial for determining the exact number and sequence of opsin genes. Each opsin gene was differentially expressed between the adult and larva, as well as between dorsal and ventral regions of adult compound eyes, highlighting the behavior, ecology, and adaptation of aquatic larva to terrestrial adult. The repertoire of opsin genes differed among dragonfly species, plausibly involved in the diversity of the habitat and behavior of each species. We also found that sex-specific yellow-red color transition in red dragonflies is regulated by redox changes in ommochrome pigments, which unveils a previously unknown molecular mechanism underlying body color change in animals. Establishment of the methods of gene functional analyses in dragonflies is desired for future studies.
The original version of this chapter was revised. An erratum to this chapter can be found at https://doi.org/10.1007/978-981-10-4956-9_18
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Like butterflies, dragonflies (including damselflies, Insecta: Odonata) are one of the most colorful insects, and their color patterns have been focused from ecological and evolutionary aspects for a long time (Tillyard 1917; Corbet 1999; Bybee et al. 2016). Dragonflies are well-known insects, and almost all Japanese people know the songs Aka-tombo (= red dragonflies; a symbol of autumn in Japan) and Tombo-no-megane (= eye glasses of dragonflies; a metaphor for the colorful compound eyes of dragonflies) (Ueda 2004; Inoue and Tani 2010). Despite the fact that the detailed genetic analyses of pattern formation in butterfly adult wing and larval body have progressed greatly in recent years as described in this book, the molecular biological study of dragonfly’s color pattern formation has just started.
2 Important Role of Color Pattern for Partner Recognition in Dragonflies
Unlike most insects, drastic adult color transitions are widely recognized among dragonflies, resulting in conspicuous sexual dimorphism. In red dragonfly species, body colors of males turn from yellow to red in the course of sexual maturation, whereas females are yellowish throughout their adult lives in general (Fig. 17.1a–c). Moreover, many dragonfly species have color polymorphism even in the same sex (Fig. 17.1c–f, i–n), which is genetically controlled at least in several species (Futahashi 2016a). Considering that gynandromorph specimens display discontinuous male/female mosaicism in their coloration (Fig. 17.1g, h), sex-specific color formation is regulated cell-autonomously in dragonflies.
Intraspecific color pattern diversity in dragonflies. (a–d) Sexual dimorphism, male color transition, and female color polymorphism of Sympetrum cordulegaster. (e–f) Female color polymorphism of Ischnura senegalensis. Arrow indicates a blue spot existed in males and androchrome females. (g) Gynandromorph of I. senegalensis showing the main region with female coloration and the posterior left side (arrow) with male coloration and appendage (Photo courtesy of Mitsutoshi Sugimura). (h) Gynandromorph of Crocothemis servilia showing the main region with male coloration and the anterior right side with female coloration (Photo courtesy of Kohji Tanaka). (i–n) Wing color polymorphism of Mnais species. Territorial males have orange (i) or brown (l) wings, whereas female mimicking males have hyaline (j) or pale orange (m) wings. Females have hyaline (k) or pale orange (n) wings. (i–k, m, n) M. costalis. (l) M. pruinosa (See also Fig. 17.3)
In many dragonfly species, adult body color plays important roles in partner recognition (Corbet 1999; Svensson et al. 2007; Córdoba-Aguilar 2008; Svensson et al. 2014; Takahashi et al. 2014; Beatty et al. 2015; Drury et al. 2015). Interspecific or male-male connection has sometimes been observed in the field between similar-colored individuals (Fig. 17.2a–c), and interspecific hybrids has been reported occasionally (Fig. 17.2d–f) (Corbet 1999; Futahashi 1999; Futahashi and Futahashi 2007; Moriyasu and Sugimura 2007; Ozono et al. 2012; Sánchez-Guillén et al. 2014; Futahashi 2016a). Parent combination of hybrid specimen can be determined by biparentally inherited nuclear DNA and maternally inherited mitochondrial DNA analyses (Fig. 17.2g), and it has been reported that males of Sympetrum eroticum are apt to catch females of other species (Futahashi 1999; Futahashi and Hayashi 2004a), suggesting that the direction of gene flow with hybridization is nonreciprocal in some cases. These misidentifications in dragonflies may be attributed to their poor sense of audition and olfaction; dragonflies lack the auditory organs, and their antennae are less developed (Yager 1999; Cocroft and Rodríguez 2005).
Interspecific copulation and hybrid of dragonflies. (a) Normal copulation of Sympetrum croceolum. (b) Interspecific copulation between S. croceolum male and Sympetrum speciosum female (Figure modified from Ozono et al. 2012). (c) Normal copulation of S. speciosum. (d) Male of S. croceolum. (e) Interspecific hybrid male from S. croceolum male and S. speciosum female. (f) Male of S. speciosum. (g) Nuclear and mitochondrial DNA analyses of S. croceolum, S. speciosum, and interspecific hybrid between S. croceolum male and S. speciosum female. The internal transcribed spacer 1 (ITS1) or cytochrome c oxidase subunit I (COI) region were used for nuclear or mitochondrial DNA marker, respectively. Arrows indicate species specific nucleotides
3 Wing Color Polymorphism and Presumptive Character Displacement in Japanese Mnais Species
In order to avoid interspecific mating or aggression, presumptive character displacement has been reported in some species, wherein interspecific color differences are larger in sympatric populations than in allopatric populations (Waage 1975; Suzuki 1984; Tynkkynen et al. 2004; Hayashi et al. 2004b; Hassall 2014; Drury and Grether 2014; Tsubaki and Okuyama 2016). Here I introduce an interesting example of wing color polymorphism in the two closely related Mnais species, M. costalis and M. pruinosa, in Japan (Hayashi et al. 2004a, b; Ozono et al. 2012). These two species can be distinguished by nuclear ITS1 sequences, relative length of wing to head in adult males, shape of adult wing pterostigma, and the shape of larval caudal gill (Hayashi et al. 2004a, b). On the other hand, interspecific hybrids have been discovered occasionally and multiregional introgression of mitochondrial DNA is recognized between these two species (Hayashi et al. 2004a, 2005; Futahashi and Hayashi 2004b). Both species exhibit complex wing color polymorphism (Figs. 17.1i–n and 17.3), in which orange-winged males, hyaline-winged males, and hyaline-winged females appear widely in Japan (Fig. 17.1i–k, Asahina 1976; Hayashi et al. 2004b; Ozono et al. 2012). Male orange/hyaline wing polymorphism of M. costalis can be explained by a single autosomal locus, and the orange-winged phenotype is dominant (Tsubaki 2003). Previous ecological studies have shown that orange-winged males are territorial, whereas hyaline-winged males are female-mimics and usually non-territorial sneakers (Nomakuchi et al. 1984; Tsubaki et al. 1997; Hayashi et al. 2004b). In addition to these three major phenotypes, the following three phenotypes are recognized in some populations: brown-winged males of M. pruinosa, pale orange-winged males and females of M. costalis (Fig. 17.1l–n, Asahina 1976; Hayashi et al. 2004b; Ozono et al. 2012). Thus, in M. costalis, there are three (orange, pale orange, and hyaline) and two (pale orange and hyaline) wing color forms for males and females, respectively, whereas in M. pruinosa, three (brown, orange, and hyaline) and one (hyaline) wing color forms exist in males and females, respectively. Interestingly, wing polymorphism is associated with abdominal body coloration: whitish in mature territorial males (orange or brown wings) (Fig. 17.1i, l) and metallic green in female-mimicking males and females (hyaline or pale orange wings) (Fig. 17.1j, k, m, n, Asahina 1976; Hayashi et al. 2004b; Ozono et al. 2012). Exceptional untransparent white-winged phenotypes have been reported in the Boso Peninsula population of M. pruinosa (Asahina 1976), although these white-winged phenotypes are now almost extinct (Futahashi and Hayashi 2004b; Ozono et al. 2012).
Wing color polymorphism of two Japanese Mnais species. (a) Geographical variation of M. costalis and M. pruinosa. (b) Hypothetical evolutionary model of wing color polymorphism. The photos of each form are shown in Fig. 17.1i–n. As described Fig. 17.3a, wing color polymorphism varied among populations in sympatric region. Pale orange-winged males and females often appear together with hyaline-winged males and females, respectively. For example, three male forms (orange, pale orange, and hyaline) and two female forms (pale orange and hyaline) emerge simultaneously in some populations of M. costalis. Figure modified from Hayashi et al. 2004b; Ozono et al. 2012
Geographic variation of wing color polymorphism is associated with cohabitation (Suzuki 1984; Hayashi et al. 2004b; Tsubaki and Okuyama 2016). In allopatric regions, males of both M. costalis and M. pruinosa show orange/hyaline wing color polymorphism, whereas females are all hyaline-winged and monomorphic in both species (Fig. 17.3a). In central Japan where both species cohabit, males of M. costalis show only orange wings, while males of M. pruinosa show only hyaline wings in general (Fig. 17.3a). In M. costalis, pale orange-winged females appear prominently in the southern area, where both males and females can be distinguished solely by wing coloration (Fig. 17.3a). In addition to wing color polymorphism, sympatric populations of M. costalis show larger body size and prefer sunnier habitats than M. pruinosa and allopatric populations of M. costalis, suggesting that multiple character displacements emerged in central Japan to avoid interspecific mating (Tsubaki and Okuyama 2016). In western Japan, however, males of both species are polymorphic even in sympatric regions. It should be noted that two forms of female-mimicking males (pale orange and hyaline) appear in accordance with female color polymorphism in M. costalis (Fig. 17.3). In the southwestern area, territorial males of M. pruinosa have brown wings instead of orange, where both males and females can be distinguished solely by wing coloration (Fig. 17.3b, rightmost). Although it is not clear why wing color polymorphism is maintained in western Japan even in sympatric region, it has been reported that degrees of mitochondrial introgression are smaller in western Japan than in central Japan (Hayashi et al. 2005), suggesting that reproductive isolating mechanism between the two Mnais species is more robust in western Japan. Hayashi et al. (2004b) proposed the evolutionary model of Mnais wing polymorphism, in which the following three stages are hypothesized (Fig. 17.3b):
-
1.
In allopatric populations (eastern or southeastern Japan), males exhibit orange (territorial) and hyaline (female-mimicking) wing color polymorphism, while females are monomorphic (hyaline) in both species.
-
2.
In the early stage of cohabitation (central Japan), males become monomorphic (only orange in M. costalis and only hyaline in M. pruinosa), and pale orange-winged females of M. costalis emerge in some places whereas all females are hyaline-winged in M. pruinosa. It should be noted that hyaline-winged males of M. pruinosa in central Japan show whitish abdomen like territorial males and have flexible territorial strategy (Fig. 17.3b middle, Siva-Jothy and Tsubaki 1989; Hayashi et al. 2004b).
-
3.
In the late stage of cohabitation (western Japan), both territorial (orange or brown) and female-mimicking males (hyaline or pale orange) appear in both species once again.
According to this scenario, pale orange-winged males and brown males are likely to appear secondarily. The brown/orange color difference may have occurred because the wing color was lost in early sympatry and was reconstructed without models.
Orange wing color of M. costalis is derived from tyrosine, suggesting that pigments of orange wing are kinds of melanin (Hooper et al. 1999). Considering that dopamine, a melanin precursor, is also known as a neurotransmitter, genes involved in melanin synthesis pathway are strong candidates for analysis of pleiotropic effects on wing color formation and territoriality. Genetic mechanisms underlying wing color polymorphism deserve future studies.
4 Identification of Remarkable Number of Opsin Genes in Dragonflies
Because dragonflies visually recognize environment, foods, enemies, rivals, and mates, their sense of vision has been studied using electrophysiological approach. Critical flicker frequency test has revealed that dragonflies can discriminate beyond 300 Hz, suggesting that they have keen dynamic vision (McFarland and Lowe 1983). Meanwhile, based on anatomical studies, it has been hypothesized that dragonflies have approximately 20/2000 vision (Kirschfeld 1976). In addition to temporal and spatial resolution, wavelength discrimination capability (i.e., color vision) of dragonflies has been investigated, and previous studies have shown that they have three to five classes of photoreceptors (Autrum and Kolb 1968; Eguchi 1971; Meinertzhagen et al. 1983; Yang and Osorio 1991; Bybee et al. 2012; Huang et al. 2014).
Evolution of animal color vision is strongly correlated with the diversity of opsin genes (Briscoe and Chittka 2001; Terakita 2005; Briscoe 2008, Shichida and Matsuyama 2009; Hering et al. 2012; Cronin et al. 2014). Specific types of opsin gene produce light sensors sensitive to specific wavelength light. For example, human beings possess three opsin genes sensitive to blue, green, or red light and can perceive light from purple to red but not ultraviolet (UV). On the other hand, honey bees and fruit flies possess an opsin gene for UV light but not for red light, which allows them to recognize UV light, instead of discriminating red from gray. It has been thought that 2–5 opsin proteins are involved in color vision in most animals (Cronin et al. 2014).
Recently, we discovered that dragonflies possess surprisingly many opsin genes by RNA sequencing (RNA-seq) analyses using adult and larval visual organs (Futahashi et al. 2015). First we surveyed the visual transcriptomics of the red dragonfly Sympetrum frequens (Libellulidae). After de novo assembly using Trinity software, we obtained 60 contigs with high similarity to insect opsin proteins. When we aligned these contigs, many of them seemed to be partial or chimeric (gray and blue arrows in Fig. 17.4a). We also obtained 144 opsin gene-like contigs from the white-tailed skimmer dragonfly Orthetrum albistylum (Libellulidae) and found that chimeric pattern was different between these two species (Fig. 17.4a). We often encountered similar problem of chimeric contigs in de novo assembly when we focused on paralogous genes. To overcome this problem, we carefully checked and manually corrected each of the contig sequences using Integrative Genomics Viewer (Thorvaldsdóttir et al. 2013) (Fig. 17.4b). Through this manual correction, we also found that partial sequence information is often lost in automatically assembled contigs among highly paralogous genes, due to merging of several similar sequences into one (Futahashi 2016b). We verified the revised sequences by RT-PCR and DNA sequencing. Consequently, we obtained the presumably full length sequences of 20 opsin genes, consisting of 4 nonvisual opsin genes and 16 visual opsin genes of 1 UV, 5 short wavelength (SW), and 10 long wavelength (LW) type from both S. frequens and O. albistylum (Fig. 17.4c). Next we inspected the draft genome data of the scarce chaser dragonfly Ladona fulva (Libellulidae) and identified the same set of 20 opsin genes (Fig. 17.4c). No other opsin genes could be found in the genome. Molecular phylogenetic analysis revealed that the 20 opsin genes of these three species formed distinct 20 monophyletic clusters (Fig. 17.4c), indicating that the common ancestor of the libellulid dragonflies had these 20 genes.
Identification and manual assembly of 20 opsin genes in three libellulid dragonflies. (a) Results of de novo assembly by Trinity before manual correction. LW opsin gene-like contigs were aligned based on the similarity of ninaE/Rh1 gene of Drosophila melanogaster. The presumptive seven transmembrane regions are shaded by red. Blue and gray arrows indicate chimeric and partial contigs, respectively. (b) Manual correction of the contig sequences using Integrative Genomics Viewer. Cyan and sky-blue colors mean paired end reads mapped on the different contigs. (c) Molecular phylogeny of 20 opsin genes of three libellulid dragonflies inferred from 795 aligned amino acid sites. On each node, bootstrap values are indicated in the order of neighbor-joining method/maximum-likelihood method. Accession numbers are shown in parentheses. On the genome of L. fulva, seven LW opsin genes (LWC1, LWD1, LWE1, and LWF1–F4) and three SW opsin genes (SWc1-c3) were located in tandem, respectively (Figure modified from Futahashi 2016b)
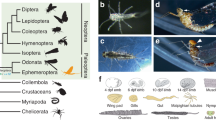
Opsin genes of dragonflies are extraordinarily large in number compared with other insects (Fig. 17.5a). Why do dragonflies have so many opsin genes? In dragonflies, the structure and function of compound eyes are markedly different between not only adult and larva but also dorsal and ventral regions of adult eyes (Fig. 17.5b–c) (Labhart and Nilsson 1995). Electrophysiological analysis of S. frequens revealed that the dorsal eye region was sensitive to a short wavelength range from UV (300 nm) to blue-green light (500 nm), whereas the ventral eye region was sensitive to a broader wavelength range from UV to red light (620 nm) (Fig. 17.5d). Interestingly, most of the opsin genes were expressed only at a specific life stage and in a specific region (Fig. 17.6). Although many opsin genes were expressed in adults, relatively small number of opsin genes was expressed in larvae, reflecting their lifestyle under water with less visual dependence. In adult compound eyes, most SW and LW opsin genes were, respectively, expressed in the dorsal and ventral regions in accordance with their spectral sensitivity, reflecting that dorsal eyes mainly perceive the SW-rich light directly from the sky, whereas the ventral eyes perceive reflected light from objects on the ground.
Insect opsin genes and spectral sensitivity of adult compound eyes of Sympetrum frequens. (a) Numbers of opsin genes of ultraviolet type (UV), short wavelength type (SW), long wavelength type (LW), rhodopsin7-like (Rh7), arthropsin type (arth), pteropsin type (ptero), and retinal G protein-coupled receptor-like (RGR) are mapped on the insect phylogeny (Misof et al. 2014). (b) Frontal view of adult head of S. frequens. (c) Larva of S. frequens (Photo courtesy of Akira Ozono). (d) Spectral sensitivity of the dorsal and ventral regions of adult eyes of S. frequens measured by electroretinography (Figure modified from Futahashi et al. 2015)
Numbers and expression patterns of each type of opsin gene of 12 dragonfly species. Phylogenetic relationship of the dragonflies (Futahashi 2014) is shown on the top: S.fre., Sympetrum frequens; O.alb., Orthetrum albistylum; S.uchi., Somatochlora uchidai; M.amp., Macromia amphigena; A.sie., Anotogaster sieboldii; T.pry., Tanypteryx pryeri; A.mel., Asiagomphus melaenops; A.par., Anax parthenope; E.sup., Epiophlebia superstes; I.asi., Ischnura asiatica; M.cos., Mnais costalis; I.per., Indolestes peregrinus. SW and LW opsin genes are categorized into three (a–c) and six (A–F) groups, respectively. In expression pattern, major tissues and stages expressing each group of opsin genes are shown in parentheses, wherein D, V, O, and L indicate dorsal region of adult eyes, ventral region of adult eyes, adult head region containing ocelli, and larval whole head, respectively (Figure modified from Futahashi et al. 2015)
5 Diversity of Opsin Genes among Dragonflies
Body and wing color pattern, behavior, and microhabitats of dragonflies are variable among the families (Corbet 1999; Ozono et al. 2012). To investigate the opsin gene repertoire across dragonflies, comparative RNA-seq analyses were performed in additional 10 species representing 10 different dragonfly families: Somatochlora uchidai (Corduliidae), Macromia amphigena (Macromiidae), Anotogaster sieboldii (Cordulegastridae), Tanypteryx pryeri (Petaluridae), Asiagomphus melaenops (Gomphidae), Anax parthenope (Aeshnidae), Epiophlebia superstes (Epiophlebiidae), Ischnura asiatica (Coenagrionidae), Mnais costalis (Calopterygidae), and Indolestes peregrinus (Lestidae) (Fig. 17.6) (Futahashi et al. 2015). The former six species belong to true dragonflies (suborder Anisoptera), while the latter three species belong to damselflies (suborder Zygoptera). E. superstes belongs to ancient dragonflies (suborder Anisozygoptera, sometimes including into Anisoptera) (Ozono et al. 2012; Futahashi 2014). Among dragonfly families, the total number of opsin genes varied widely from 15 to 33 (Fig. 17.6).
One of the significant advantages of RNA-seq analyses is that the gene expression information of different developmental stages in multiple species could be efficiently obtained. Based on molecular phylogeny and expression pattern, SW and LW opsin genes were categorized into three (a, b, and c) and six (A, B, C, D, E, and F) groups, respectively (Fig. 17.6). Nonvisual opsin genes were scarcely expressed in the larval and adult visual organs of all examined species (Futahashi et al. 2015). Stage- and region-specific expressions of opsin genes were widely conserved across dragonfly species as follows:
-
1.
The group-a SW, group-B LW, and group-C LW opsin genes were predominantly expressed in larvae.
-
2.
The group-b SW and group-F LW opsin genes were mainly expressed in the ventral region of adult compound eyes.
-
3.
The group-c SW and group-E LW opsin genes were primarily expressed in the dorsal region of adult compound eyes.
-
4.
The group-D LW opsin genes were specifically expressed in the adult ocelli (Fig. 17.6).
The dorsoventrally differentiated expression patterns were obscure in the three damselfly species (Fig. 17.6). It should be noted that compensational expression patterns associated with losses of some visual opsin genes were observed (e.g., loss of the ocellus-specific group-D LW opsin gene entailed ocellus-associated expression of the group-C or group-E genes (Fig. 17.6)). Given that the group -C, -D, -E, and -F genes were located in tandem on the genome of L. fulva, the rearrangement among these genes may have occurred in the course of evolution, resulting in lineage-specific expression pattern changes of these genes. Thus, dragonflies may utilize different sets of opsin genes depending on types of light environment, which can be achieved by an extraordinary increase in the number of opsin genes.
The repertoire of opsin genes differed among dragonfly species, suggesting that the opsin genes may have evolved according to the habitat or behavior of each species. For example, the absence of the SW opsin genes at larval stage coincided with their sand- or pit-dwelling behaviors in A. sieboldii, T. pryeri, and A. melaenops, whereas the multitude of SW and/or LW opsin gene numbers in the dorsal region of adult compound eyes are correlated with twilight flying activity for predation in S. uchidai, M. amphigena, A. parthenope, and E. superstes (Fig. 17.6; Futahashi et al. 2015). Plausibly, although speculative, the large variation of opsin genes is associated with the evolution of diverse color pattern in dragonflies.
The variety and beauty of color pattern are also prominent in Lepidopteran and Coleopteran insects, although they have only a few opsin genes (Fig. 17.5a). The small numbers of opsin genes in these insects may be attributed to nocturnal lifestyle of their ancestors like mammals (Briscoe and Chittka 2001; Feuda et al. 2016). By contrast, almost all dragonfly species are diurnal, and they diverged from other insects over 350 million years ago (Fig. 17.5a, Misof et al. 2014).
6 Molecular Mechanisms Underlying Color Changes in Red Dragonflies
Dragonflies display a wide variety of coloration such as red, yellow, blue, and green. Most of animal colors are derived from structural colors and/or pigment colors. The mechanisms of structural coloration of several dragonfly species have been recently investigated, wherein multilayer structures are generally involved in iridescent coloration (Vukusic et al. 2004; Hariyama et al. 2005; Schultz and Fincke 2009; Stavenga et al. 2012; Nixon et al. 2013, 2015; Guillermo-Ferreira et al. 2015). Non-iridescent blue color is also structural, attributed to coherent light scattering from the quasi-ordered nanostructures within pigment cells (Prum et al. 2004). By contrast, information on pigments in dragonflies is still limited.
We analyzed the red epidermal pigments from three species of red dragonfly, namely, the autumn darter Sympetrum frequens, the summer darter Sympetrum darwinianum, and the scarlet skimmer Crocothemis servilia (Futahashi et al. 2012). Two ommochrome pigments, xanthommatin (vivid red color in reduced form) and decarboxylated xanthommatin (dull red color in reduced form) were consistently identified in all these species (Fig. 17.7a), in which the ratio of xanthommatin is higher in vivid red color species. Previous studies have shown that the color of ommochrome pigments changes reversibly by redox reactions in vitro (Linzen 1974). By injecting a reductant (vitamin C) solution, we confirmed that the yellowish body color of both immature males and mature females changed into red as observed in mature males (Fig. 17.7b). Redox conditions of the extracted ommochrome pigments were measured electrochemically, and the relative abundance of the oxidized and reduced forms of pigments were evaluated. In all three species, only the mature males exhibited very high proportions of the reduced ommochrome pigments (Fig. 17.7c), indicating that sex-specific color change in mature red dragonflies is primarily attributed to redox states of the ommochrome pigments (Futahashi et al. 2012).
Pigment-based color changes in animals are mainly attributed to the following three mechanisms: synthesis and degradation of pigments, changes in localization of pigments, and accumulation of pigments from food (Stevens and Merilaita 2011). Red dragonflies adopt a previously unknown mechanism, namely, a body color change by redox reaction of the pigments. Male-specific color change of dragonflies has been considered as an ecologically important trait for reproductive success. Considering that mature males exhibit territorial behavior under the scorching sun and the reduced pigments show antioxidant abilities (Futahashi et al. 2012), male-specific red pigments may have additional role in preventing oxidative stress from UV radiation.
Redox-dependent color change of the ommochrome pigments in red dragonflies. (a) Chromatograms of ommochrome pigments from males of three red dragonflies. Blue lines denote the acetonitrile gradient. (b) Reductant-induced yellow/red color change. Arrows indicate the injection sites. (c) Reduced form ratios of the extracted ommochrome pigments. Means and standard deviation are shown (n = 10 ~ 12) (Figure modified from Futahashi et al. 2012)
7 Conclusion and Perspective
The well-developed sense of sight and the great variety of color pattern in dragonflies have been already pointed out a century ago (Tillyard 1917). Recent progress on molecular mechanisms of color vision and color formation unveiled the outstanding diversity of visual opsin genes and the unique mechanisms of body color changes in dragonflies. Meanwhile, only limited information is available on genes potentially involved in color formation (Chauhan et al. 2014, 2016). Rapid spread of next-generation sequencing technology makes it easier than ever to analyze non-model organisms, although careful evaluation for de novo assembly is still important as described above, especially without genomic information. Molecular bases underlying color pattern formation and its evolution in dragonflies are just as fascinating and challenging as in butterflies. Recently, effective RNAi- and genome-editing methods have been developed for gene functional analyses in butterflies (Ando and Fujiwara 2013; Nishikawa et al. 2015; Li et al. 2015; Perry et al. 2016; Zhang and Reed 2016; Beldade and Peralta 2017). Applying these methods to dragonflies will be an important step toward future studies in this field (Okude et al. 2017).
References
Ando T, Fujiwara H (2013) Electroporation-mediated somatic transgenesis for rapid functional analysis in insects. Development 140:454–458
Asahina S (1976) A revisional study of the genus Mnais (Odonata, Calopterygidae) VIII. A proposed taxonomy of Japanese Mnais. Tombo 19:2–16
Autrum H, Kolb G (1968) Spektrale Empfindlichkeit einzelner Sehzellen der Aeschniden. Z Vgl Physiol 60:450–477
Beatty CD, Andrés JA, Sherratt TN (2015) Conspicuous coloration in males of the damselfly Nehalennia irene (Zygoptera: Coenagrionidae): do males signal their unprofitability to other males? PLoS One 10:e0142684
Beldade P, Peralta CM (2017) Developmental and evolutionary mechanisms shaping butterfly eyespots. Curr Opin Insect Sci 19:22–29
Briscoe AD (2008) Reconstructing the ancestral butterfly eye: focus on the opsins. J Exp Biol 211:1805–1813
Briscoe AD, Chittka L (2001) The evolution of color vision in insects. Annu Rev Entomol 46:471–510
Bybee SM, Johnson KK, Gering EJ, Whiting MF, Crandall KA (2012) All the better to see you with: a review of odonate color vision with transcriptomic insight into the odonate eye. Org Divers Evol 12:241–250
Bybee S, Córdoba-Aguilar A, Duryea MC, Futahashi R, Hansson B, Lorenzo-Carballa MO, Schilder R, Stoks R, Suvorov A, Svensson EI, Swaegers J, Takahashi Y, Watts PC, Wellenreuther M (2016) Odonata (dragonflies and damselflies) as a bridge between ecology and evolutionary genomics. Front Zool 13:46
Chauhan P, Hansson B, Kraaijeveld K, de Knijff P, Svensson EI, Wellenreuther M (2014) De novo transcriptome of Ischnura elegans provides insights into sensory biology, colour and vision genes. BMC Genomics 15:808
Chauhan P, Wellenreuther M, Hansson B (2016) Transcriptome profiling in the damselfly Ischnura elegans identifies genes with sex-biased expression. BMC Genomics 17(1):985
Cocroft RB, Rodríguez RL (2005) The behavioral ecology of insect vibrational communication. Bioscience 55:323–334
Corbet PS (1999) Dragonflies, behavior and ecology of odonata. Cornell University Press
Córdoba-Aguilar A (2008) Dragonflies and damselflies: model organisms for ecological and evolutionary research. Oxford, Oxford University Press
Cronin TW, Johnsen S, Marshall NJ, Warrant EJ (2014) Visual ecology. PrincetonUniversity Press, Princeton
Drury JP, Grether GF (2014) Interspecific aggression, not interspecific mating, drives character displacement in the wing coloration of male rubyspot damselflies (Hetaerina). Proc Biol Sci 281:20141737
Drury JP, Anderson CN, Grether GF (2015) Seasonal polyphenism in wing coloration affects species recognition in rubyspot damselflies (Hetaerina spp.) J Evol Biol 28:1439–1452
Eguchi E (1971) Fine structure and spectral sensitivities of retinular cells in the dorsal sector of compound eyes in the dragonfly, Aeschna. Z Vgl Physiol 71:201–218
Feuda R, Marlétaz F, Bentley MA, Holland PW (2016) Conservation, duplication, and divergence of five opsin genes in insect evolution. Genome Biol Evol 8:579–587
Futahashi R (1999) Notes on unusual connection and copulation in some species of dragonflies. Aeschna 36:47–55
Futahashi R (2014) A revisional study of Japanese dragonflies based on DNA analysis (2). Tombo 56:57–59
Futahashi R (2016a) Color vision and color formation in dragonflies. Curr Opin Insect Sci 17:32–39
Futahashi R (2016b) RNAseq analyses and opsin genes of dragonflies: the importance of manual assembly. Sanshi-Konchu Biotec 85(1):13–18
Futahashi R, Futahashi H (2007) A record of a black mutant of Nannophya pygmaea Rambur, 1842. Tombo 50:73–74
Futahashi R, Hayashi F (2004a) DNA analysis of hybrids between Sympetrum eroticum eroticum and S. baccha matutinum. Tombo 47:31–36
Futahashi R, Hayashi F (2004b) Distribution patterns of two damselfly species, Mnais costalis and M. strigata, in the Boso Peninsula, Chiba Prefecture. Tombo 47:41–46
Futahashi R, Kurita R, Mano H, Fukatsu T (2012) Redox alters yellow dragonflies into red. Proc Natl Acad Sci U S A 109:12626–12631
Futahashi R, Kawahara-Miki R, Kinoshita M, Yoshitake K, Yajima S, Arikawa K, Fukatsu T (2015) Extraordinary diversity of visual opsin genes in dragonflies. Proc Natl Acad Sci U S A 112:E1247–E1256
Guillermo-Ferreira R, Bispo PC, Appel E, Kovalev A, Gorb SN (2015) Mechanism of the wing colouration in the dragonfly Zenithoptera lanei (Odonata: Libellulidae) and its role in intraspecific communication. J Insect Physiol 81:129–136
Hariyama T, Hironaka M, Horiguchi H, Stavenga DG (2005) The leaf beetle, the jewel beetle, and the damselfly; insects with a multilayered show case. In: Shimozawa T, Hariyama T (eds) Structural colors in biological systems: principles and applications. University Press, Osaka, pp 153–176
Hassall C (2014) Continental variation in wing pigmentation in Calopteryx damselflies is related to the presence of heterospecifics. PeerJ 2:e438
Hayashi F, Dobata S, Futahashi R (2004a) Macro- and microscale distribution patterns of two closely related Japanese Mnais species inferred from nuclear ribosomal DNA, ITS sequences and morphology (Zygoptera: Odonata). Odonatologica 33(4):399–412
Hayashi F, Dobata S, Futahashi R (2004b) A new approach to resolve the taxonomic and ecological problems of Japanese Mnais damselflies (Odonata: Calopterygidae) (1). General remarks Aeschna 41:1–14
Hayashi F, Dobata S, Futahashi R (2005) Disturbed population genetics: suspected introgressive hybridization between two Mnais damselfly species (Odonata). Zool Sci 22(8):869–881
Hering L, Henze MJ, Kohler M, Kelber A, Bleidorn C, Leschke M, Nickel B, Meyer M, Kircher M, Sunnucks P, Mayer G (2012) Opsins in Onychophora (velvet worms) suggest a single origin and subsequent diversification of visual pigments in arthropods. Mol Biol Evol 29(11):3451–3458
Hooper RE, Tsubaki Y, Siva-Jothy MT (1999) Expression of a costly, plastic secondary sexual trait is correlated with age and condition in a damselfly with two male morphs. Physiol Entomol 24:364–369
Huang SC, Chiou TH, Marshall J, Reinhard J (2014) Spectral sensitivities and color signals in a polymorphic damselfly. PLoS One 9:e87972
Inoue K, Tani K (2010) All about red dragonflies. Tombow, Osaka
Kirschfeld K (1976) The resolution of lens and compound eyes. In: Neural principles in vision. Springer, Berlin, pp 354–370
Labhart T, Nilsson DE (1995) The dorsal eye of the dragonfly Sympetrum: specializations for prey detection against the blue sky. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 1995(176):437–453
Li X, Fan D, Zhang W, Liu G, Zhang L, Zhao L, Fang X, Chen L, Dong Y, Chen Y, Ding Y, Zhao R, Feng M, Zhu Y, Feng Y, Jiang X, Zhu D, Xiang H, Feng X, Li S, Wang J, Zhang G, Kronforst MR, Wang W (2015) Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat Commun 6:8212
Linzen B (1974) The tryptophan to ommochrome pathway in insects. Adv Insect Physiol 10:117–246
McFarland WN, Lowe ER (1983) Wave produced changes in underwater light and their relations to vision. Environ Biol Fish 8:173–184
Meinertzhagen IA, Menzel R, Kahle G (1983) The identification of spectral receptor types in the retina and lamina of the dragonfly Sympetrum rubicundulum. J Comp Physiol 151:295–310
Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, Niehuis O, Petersen M, Izquierdo-Carrasco F, Wappler T, Rust J, Aberer AJ, Aspöck U, Aspöck H, Bartel D, Blanke A, Berger S, Böhm A, Buckley TR, Calcott B, Chen J, Friedrich F, Fukui M, Fujita M, Greve C, Grobe P, Gu S, Huang Y, Jermiin LS, Kawahara AY, Krogmann L, Kubiak M, Lanfear R, Letsch H, Li Y, Li Z, Li J, Lu H, Machida R, Mashimo Y, Kapli P, McKenna DD, Meng G, Nakagaki Y, Navarrete-Heredia JL, Ott M, Ou Y, Pass G, Podsiadlowski L, Pohl H, von Reumont BM, Schütte K, Sekiya K, Shimizu S, Slipinski A, Stamatakis A, Song W, Su X, Szucsich NU, Tan M, Tan X, Tang M, Tang J, Timelthaler G, Tomizuka S, Trautwein M, Tong X, Uchifune T, Walzl MG, Wiegmann BM, Wilbrandt J, Wipfler B, Wong TK, Wu Q, Wu G, Xie Y, Yang S, Yang Q, Yeates DK, Yoshizawa K, Zhang Q, Zhang R, Zhang W, Zhang Y, Zhao J, Zhou C, Zhou L, Ziesmann T, Zou S, Li Y, Xu X, Zhang Y, Yang H, Wang J, Wang J, Kjer KM, Zhou X (2014) Phylogenomics resolves the timing and pattern of insect evolution. Science 346(6210):763–767
Moriyasu A, Sugimura M (2007) An interspecific hybrid between Sympetrum croceolum and S. speciosum. Mon J Entomol 433:44–45
Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, Ando T, Suzuki Y, Sugano S, Fujiyama A, Kosugi S, Hirakawa H, Tabata S, Ozaki K, Morimoto H, Ihara K, Obara M, Hori H, Itoh T, Fujiwara H (2015) A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat Genet 47(4):405–409
Nixon MR, Orr AG, Vukusic P (2013) Subtle design changes control the difference in colour reflection from the dorsal and ventral wing-membrane surfaces of the damselfly Matronoides cyaneipennis. Opt Express 21:1479–1488
Nixon MR, Orr AG, Vukusic P (2015) Wrinkles enhance the diffuse reflection from the dragonfly Rhyothemis resplendens. J R Soc Interface 12:20140749
Nomakuchi S, Higashi K, Harada M, Maeda M (1984) An experimental study of the territoriality in Mnais pruinosa pruinosa Selys (Zygoptera: Calopterygidae). Odonatologica 13:259–267
Okude G, Futahashi R, Kawahara-Miki R, Yoshitake K, Yajima S, Fukatsu T (2017) Electroporation-mediated RNA interference reveals a role of multicopper oxidase 2 gene in dragonfly's cuticular pigmentation. Appl Entomol Zool. doi:10.1007/s13355-017-0489-9
Ozono A, Kawashima I, Futahashi R (2012) Dragonflies of Japan, Bunichi-Sogo Syuppan., Co. Ltd
Perry M, Kinoshita M, Saldi G, Huo L, Arikawa K, Desplan C (2016) Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Nature 535(7611):280–284
Prum RO, Cole JA, Torres RH (2004) Blue integumentary structural colours in dragonflies (Odonata) are not produced by incoherent Tyndall scattering. J Exp Biol 207:3999–4009
Sánchez-Guillén RA, Córdoba-Aguilar A, Cordero-Rivera A, Wellenreuther M (2014) Genetic divergence predicts reproductive isolation in damselflies. J Evol Biol 27:76–87
Schultz TD, Fincke OM (2009) Structural colours create a flashing cue for sexual recognition and male quality in a neotropical giant damselfly. Funct Ecol 23:724–732
Shichida Y, Matsuyama T (2009) Evolution of opsins and phototransduction. Philos Trans R Soc Lond Ser B Biol Sci 364:2881–2895
Siva-Jothy MT, Tsubaki Y (1989) Variation in copulation duration in Mnais pruinosa pruinosa Selys (Odonata: Calopterygidae). I. Alternative mate securing tactics and sperm precedence. Behav Ecol Sociobiol 24:39–45
Stavenga DG, Leertouwer HL, Hariyama T, De Raedt HA, Wilts BD (2012) Sexual dichromatism of the damselfly Calopteryx japonica caused by a melanin-chitin multilayer in the male wing veins. PLoS One 7:e49743
Stevens M, Merilaita S (2011) Animal camouflage: mechanisms and function. Cambridge University Press, Cambridge
Suzuki K (1984) Character displacement and evolution of the Japanese Mnais damselflies (Zygoptera: Calopterygidae). Odonatologica 13:287–300
Svensson EI, Karlsson K, Friberg M, Eroukhmanoff F (2007) Gender differences in species recognition and the evolution of asymmetric sexual isolation. Curr Biol 17:1943–1947
Svensson EI, Runemark A, Verzijden MN, Wellenreuther M (2014) Sex differences in developmental plasticity and canalization shape population divergence in mate preferences. Proc Biol Sci 281:20141636
Takahashi Y, Kagawa K, Svensson EI, Kawata M (2014) Evolution of increased phenotypic diversity enhances population performance by reducing sexual harassment in damselflies. Nat Commun 5:4468
Terakita A (2005) The opsins. Genome Biol 6:213
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192
Tillyard RJ (1917) The biology of dragonflies. Cambridge University Press, Cambridge
Tsubaki Y (2003) The genetic polymorphism linked to mate-securing strategies in the male damselfly Mnais costalis Selys (Odonata: Calopterygidae). Popul Ecol 45:263–266
Tsubaki H, Okuyama H (2016) Adaptive loss of color polymorphism and character displacements in sympatric Mnais damselflies. Evol Ecol 30:811
Tsubaki Y, Hooper RE, Siva-Jothy MT (1997) Differences in adult and reproductive lifespan in the two male forms of Mnais pruinosa costalis selys (Odonata: Calopterygidae). Res Popul Ecol 39:149–155
Tynkkynen K, Rantala MJ, Suhonen J (2004) Interspecific aggression and character displacement in the damselfly Calopteryx splendens. J Evol Biol 17:759–767
Ueda T (2004) How do the Japanese see dragonflies? Kyoto University Press, Kyoto
Vukusic P, Wootton RJ, Sambles JR (2004) Remarkable iridescence in the hindwings of the damselfly Neurobasis chinensis chinensis (Linnaeus) (Zygoptera: Calopterygidae). Proc Biol Sci 271:595–601
Waage JK (1975) Reproductive isolation and the potential for character displacement in the damselflies, Calopteryx maculata and C. aequabilis (Odonata: Calopterygidae). Syst Zool 24:24–36
Yager DD (1999) Structure, development, and evolution of insect auditory systems. Microsc Res Tech 47:380–400
Yang EC, Osorio D (1991) Spectral sensitivities of photoreceptors and lamina monopolar cells in the dragonfly, Hemicordulia tau. J Comp Physiol A 169:663–669
Zhang L, Reed RD (2016) Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat Commun 7:11769
Acknowledgments
I would like to thank Mitsutoshi Sugimura and Kohji Tanaka for photos of gynandromorphic dragonflies, Akira Ozono for photo of S. frequens larva, and Genta Okude and Mizuko Osanai-Futahashi for helpful comments of the manuscript. The author’s work was supported by JSPS KAKENHI Grant Numbers 23780058, 26660276, 26711021.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
This chapter is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
Copyright information
© 2017 The Author(s)
About this chapter
Cite this chapter
Futahashi, R. (2017). Molecular Mechanisms Underlying Color Vision and Color Formation in Dragonflies. In: Sekimura, T., Nijhout, H. (eds) Diversity and Evolution of Butterfly Wing Patterns. Springer, Singapore. https://doi.org/10.1007/978-981-10-4956-9_17
Download citation
DOI: https://doi.org/10.1007/978-981-10-4956-9_17
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-4955-2
Online ISBN: 978-981-10-4956-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)