Abstract
Adolescent Idiopathic Scoliosis (AIS) exhibits as an abnormal curvature of the spine in teens. Conventional radiographic assessment of scoliosis is unreliable due to the need for manual intervention from clinicians as well as high variability in images. Current methods for automatic scoliosis assessment are not robust due to reliance on segmentation or feature engineering. We propose a novel framework for automated landmark estimation for AIS assessment by leveraging the strength of our newly designed BoostNet, which creatively integrates the robust feature extraction capabilities of Convolutional Neural Networks (ConvNet) with statistical methodologies to adapt to the variability in X-ray images. In contrast to traditional ConvNets, our BoostNet introduces two novel concepts: (1) a BoostLayer for robust discriminatory feature embedding by removing outlier features, which essentially minimizes the intra-class variance of the feature space and (2) a spinal structured multi-output regression layer for compact modelling of landmark coordinate correlation. The BoostNet architecture estimates required spinal landmarks within a mean squared error (MSE) rate of 0.00068 in 431 crossvalidation images and 0.0046 in 50 test images, demonstrating its potential for robust automated scoliosis assessment in the clinical setting.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Adolescent Idiopathic Scoliosis (AIS) is an abnormal structural, lateral, rotated curvature of the spine, which arises in children at or around puberty and could potentially lead to reduced quality of life [1]. The estimated incidence of AIS is 2.5% in the general population and only 0.25% of patients will progress to a state where treatment is necessary [2]. Early detection of progression symptoms has potential positive impacts on prognosis by allowing clinicians to provide earlier treatment for limiting disease progression.
However, conventional manual measurement involves heavy intervention from clinicians in identification of required vertebrae structures, which suffers from high inter- and intra-observer variability while being time-intensive. The accuracy of measurement is often affected by many factors such as the selection of vertebrae, the bias of observer, as well as image quality. Moreover, variabilities in measurements can affect diagnosis when assessing scoliosis progression. It is therefore important to provide accurate and robust quantitative measurements for spinal curvature. The current widely adapted standard for making scoliosis diagnosis and treatment decisions is the manual measurement of Cobb angles. These angles are derived from a posterior-anterior (back to front) X-rays and measured by selecting the most tilted vertebra at the top and bottom of the spine with respect to a horizontal line [3]. It is challenging for clinicians to make accurate measurements due to the large anatomical variation and low tissue contrast of x-ray images, which results in huge variations between different clinicians. Therefore, computer assistance is necessary for making robust quantitative assessments of scoliosis.
Segmentation and Filter-Based Method for AIS Assessment. Current computer-aided methods proposed in the literature for the estimation of Cobb angles are not ideal as part of clinical scoliosis assessment. Mathematical models such as Active Contour Model [4], Customized Filter [5] and Charged-Particle Models [6] were used to localize required vertebrae in order to derive the Cobb angle from their slopes. These methods require accurate vertebrae segmentations and feature engineering, which makes them computationally expensive and susceptible to errors caused by variation in x-ray images.
Machine Learning-Based Method for AIS Assessment. Machine learning algorithms such as Support Vector Regression (SVR) [7], Random Forest Regression (RFR) [8], and Convolutional Neural Networks (ConvNet) [9, 10] have been used for various biomedical tasks, their direct application to AIS assessment suffer from the following limitations: (1) the method’s robustness and generalizability can be compromised by the presence of outliers (such as human error, imaging artifacts, etc.) in the training data [11], which usually requires a dedicated preprocessing stage and (2) the explicit dependencies between multiple outputs (landmark coordinates) are not taken into account, which is essential for enhancing discriminative learning with respect to spinal landmark locations. While [12] successfully modified the SVR to incorporate output dependencies for the detection of spinal landmarks, their method still requires suboptimal feature extraction which does not cope with image outliers.
Proposed Method. Our proposed BoostNet achieves fully automatic clinical AIS assessment through direct spinal landmark estimation. The use of landmarks is advantageous to scoliosis assessment due to the fact that a set of spinal landmarks contain a holistic representation of the spine, which are robust to variations in local image contrast. Therefore, small local deviations in spinal landmark coordinates will not affect the overall quality of the detected spinal structure compared to conventional segmentation-based methods. Figure 1 shows our proposed BoostNet architecture overcoming the limitations of conventional AIS assessment. As shown in Fig. 1, the BoostNet architecture overcomes the limitations of conventional AIS assessment by enhancing the feature space through outlier removal and improving robustness by enforcing spinal structure.
Contribution. In summary, our work contributes in the following aspects:
-
The newly proposed BoostNet architecture can automatically and efficiently locate spinal landmarks, which provides a multi-purpose framework for robust quantitative assessment of spinal curvatures.
-
The newly proposed BoostLayer endows networks with the ability to efficiently eliminate deleterious effects of outlier features and thereby improving robustness and generalizability.
-
The newly proposed spinal structured multi-output layer significantly improves regression accuracy by explicitly enforcing the dependencies between spinal landmarks.
2 Methodology
2.1 Novel BoostNet Architecture
Our novel BoostNet architecture is designed to automatically detect spinal landmarks for comprehensive AIS assessment. Our BoostNet consists of 3 parts: (1) a series of convolutional layers as feature extractors to automatically learn features from our dataset without the need for expensive and potentially suboptimal hand-crafted features, (2) a newly designed BoostLayer (Sect. 2.1), which removes the impact of deleterious outlier features, and (3) a spinal structured multi-output layer (Sect. 2.1) that acts as a prior to alleviate the impact of small dataset by capturing essential dependencies between each spinal landmark.
Conceptualized diagram of our BoostLayer module. (a) The presence of outliers in the feature space impedes robust feature embedding. (b) The BoostLayer module detects outlier features based on a statistical properties. We use an orange dashed line to represent the outlier correction stage of the BoostLayer. For the sake of brevity, we did not include the biases and activation function in the diagram. (c) After correcting outliers, the intra-class feature variance is reduced, allowing for a more robust feature embedding.
BoostLayer. As shown in Fig. 2, the BoostLayer reduces the impact of deleterious outlier features by enhancing the feature space. The sources of outliers in medical images typically include imaging artifacts, local contrast variability, and human errors, which reduces the robustness of predictive models. The BoostLayer algorithm creatively integrates statistical outlier removal methods into ConvNets in order to boost discriminative features and minimize the impact of outliers automatically during training. The BoostLayer improves discriminative learning by minimizing the intra-class variance of the feature space. Outlier features within the context of this paper is defined as values that are greater than a predetermined threshold from the mean of the feature distribution. An overview of the algorithm is shown in Algorithm 1.

The BoostLayer functions by first computing a reconstruction (R) of some input feature (x): \(R = f(x \cdot W + b_1)\cdot W^T\,+\,b_2\) where f is the relu activation function, W is the layer weights and \(W^T\) its transpose, and \(b_{1/2}\) are the bias vectors.
The element-wise reconstruction error (\(\varepsilon \)) can be defined as \(\varepsilon = (x-R)^2\). This can alternatively be seen as the variance of a feature with respect to the latent feature distribution. What we want to establish next is a threshold such that any input (features) with reconstruction error larger than the threshold is replaced by the mean of the feature in order to minimize intra-feature variance. For our experiments, we assumed a Gaussian distribution for the feature population and used a threshold of 2 standard deviations as the criterion for determining outliers.
In other words, we want to construct an enhanced feature space (\(\hat{x}\)) such that:
where \(\mu _i\) is the estimated population mean of the \(i^{th}\) feature derived through sampling and \(\sigma _i\) is the feature’s sample standard deviation.
Each feature’s population mean can be approximated by sampling across each mini-batch during training using \(\mu \,\tilde{=}\,\frac{1}{T\times M}\sum _{k}^{T}{\sum _{i}^{M}{\bar{x}_i}}\), where M is the number of mini-batches per epoch, T is the number of epochs and \(\bar{x}\) is the sample mean of a batch. For our experiments, we used a mini-batch size of 100 and trained for 100 epochs.
Finally, we transform the revised input using the layer weights such that \(\hat{y} = f(\hat{x} \cdot W + b_1)\).
Spinal Structured Multi-output Layer. The Spinal Structured Multi-Output Layer acts as a structural prior to our output landmarks, which alleviates the impact of small datasets while improving the regression accuracy. As shown in Fig. 1, the layer captures the dependency information between the spinal landmarks in the form of a Dependency Matrix (DM) S. We define S as a spinal structured DM for the output landmarks, in which adjacent spinal landmarks are represented by 1 while distant landmarks are represented by 0. For instance, since vertebrae T1 and T3 are not directly connected, we assign their dependency value as \(S[1,3]=S[3,1]=0\) while T1 and T2 are connected so their dependency was set to \(S[1,2]=S[2,1]=1\) and so on. The spinal structured multi-output layer \(f(a_i)\) is defined as:
where \(a_i=x_i\cdot W_i + b_i\), \(S_i\) is the landmark dependency matrix, \(W_i\) the weights, and \(b_i\) the bias of landmark coordinate i.
2.2 Training Algorithm
We trained the BoostNet using mini-batch stochastic gradient descent optimization with Nesterov momentum of 0.9 and a starting learning rate of 0.01. The learning rate was adaptively halved based on validation error during training in order to tune the parameters to a local minimum. We trained the model over 1000 epochs and used Early Stopping to prevent over-fitting. During training, the loss function is optimized such that \(\mathcal {L}(X,Y,\theta ) = \sum _{i}^{c}{(Y_i-F(X))^2} + \lambda \sum _{i}^{k}{|\theta _i|}\) (where c is the number of classes, Y is the ground truth landmark coordinates, F(X) is the predicted landmark coordinates, and \(\theta \) is the set of model parameters) is minimized. The model and training algorithm was implemented in Python 2.7 using the Keras Deep Learning library [13].
2.3 Dataset
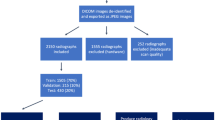
Our dataset consists of 481 spinal anterior-posterior x-ray images provided by local clinicians. All the images used for training and testing show signs of scoliosis to varying extent. Since the cervical vertebrae (vertebrae of the neck) are seldom involved in spinal deformity [14], we selected 17 vertebrae composed of the thoracic and lumbar spine for spinal shape characterization. Each vertebra is located by four landmarks with respect to four corners thus resulting in 68 points per spinal image. These landmarks were manually annotated by the authors based on visual cues. During training, the landmarks were scaled based on original image dimensions such that the range of values lies between 0–1 depending on where the landmark lies with respect to the original image (e.g. [0.5, 0.5] is exact centre of the image). We then divided our data according to 431 training/validation (Trainset) and 50 testing set (Testset) such that no patient is placed in both sets. We then trained and validated our model on the Trainset and tested the trained model on the Testset.
Data Augmentation. Since ConvNets like our BoostNet typically require large amounts of training data, we augmented our data in order teach our network the various invariance properties in our dataset. The types of augmentation used include: (a) Adding Gaussian Noise directly to our image in order to simulate inherent noise and (b) Randomly adjusting the landmark coordinates based on Gaussian distribution in order to simulate variability during data labelling.
3 Results
The BoostNet achieved superior performance in landmark detection compared to other baseline models in our crossvalidation study. Figure 3(a) shows the qualitative results of the BoostNet’s effectiveness in spinal landmark detection. The BoostNet accurately detects all the spinal landmarks despite the variations in anatomy and image contrast between different patients. The landmarks detected by the BoostNet appear to follow the general spinal curvature more closely compared to conventional ConvNet. Figure 3(b) demonstrates the effectiveness of our BoostNet in learning more discriminative features compared to an equivalent ConvNet (without BoostLayer and structured output).
Evaluation. We use the Mean Squared Error (\(MSE = E[(f(X)-Y)^2]\)) and Pearson Correlation Coefficient (\(\rho = \frac{E[f(X)]E[Y]}{\sigma _{f(X)}\sigma _{Y}}\)) between the predicted landmarks (f(X)) and annotated ground truth (Y) as the criteria of evaluating the accuracy of the estimations.
Crossvalidation. Our model achieved a reputable average MSE of 0.00068 in landmark detection based on 431 images and is demonstrated as a robust method for automatic AIS assessment. In order to validate our model as an effective way for landmark estimation, we applied a 5-fold crossvalidation of our model against the Trainset. Table 1(a) summarizes the average crossvalidation performance of our model and several baseline models including ConvNet (our model without BoostLayer and Structured Output Layer), RFR [15], and SVR [12].
Test Performance. Table 1(b) demonstrates the BoostNet’s effectiveness in a hypothetical real world setting. After training each of the models listed in the table on all 431 images from the Trainset, we evaluated each model on the Testset consisting of 50 unseen images. The BoostNet outperforms the other baseline methods based on MSE rate while showing superior qualitative results as seen in Fig. 3(a).
Analysis. The BoostNet achieved the lowest average MSE of 0.0046 and the highest correlation coefficient of 0.94 on the unseen Testset. This is due to the contributions of (1) the BoostLayer, which successfully learned robust discriminative feature embeddings as is evident in the higher accuracy in images with noticeable variability in Fig. 3(a) and (2) the spinal structured multi-output regression layer, which faithfully captured the structural information of the spinal landmark coordinates. The success of our method is further exemplified by the more than 5-fold reduction in MSE as well as more rapid convergence compared to the conventional ConvNet model Fig. 3(b).
4 Conclusion
We have proposed a novel spinal landmark estimation framework that uses our newly designed BoostNet architecture to automatically assess scoliosis. The proposed architecture creatively utilizes the feature extraction capabilities of ConvNets as well as statistical outlier detection methods to accommodate the often noisy and poorly standardized X-ray images. Intense experimental results have demonstrated that our method is a robust and accurate way for detecting spinal landmarks for AIS assessment. Our framework allows clinicians to measure spinal curvature more accurately and robustly as well as enabling researchers to develop predictive tools for measuring prospective risks based on imaging biomarkers for preventive treatment.
References
Weinstein, S.L., Dolan, L.A., Cheng, J.C., Danielsson, A., Morcuende, J.A.: Adolescent idiopathic scoliosis. Lancet 371(9623), 1527–1537 (2008)
Asher, M.A., Burton, D.C.: Adolescent idiopathic scoliosis: natural history and long term treatment effects. Scoliosis 1(1), 2 (2006)
Vrtovec, T., Pernuš, F., Likar, B.: A review of methods for quantitative evaluation of spinal curvature. Eur. Spine J. 18(5), 593–607 (2009)
Anitha, H., Prabhu, G.: Automatic quantification of spinal curvature in scoliotic radiograph using image processing. J. Med. Syst. 36(3), 1943–1951 (2012)
Anitha, H., Karunakar, A., Dinesh, K.: Automatic extraction of vertebral endplates from scoliotic radiographs using customized filter. Biomed. Eng. Lett. 4(2), 158–165 (2014)
Sardjono, T.A., Wilkinson, M.H., Veldhuizen, A.G., van Ooijen, P.M., Purnama, K.E., Verkerke, G.J.: Automatic cobb angle determination from radiographic images. Spine 38(20), 1256–1262 (2013)
Sánchez-Fernández, M., de Prado-Cumplido, M., Arenas-García, J., Pérez-Cruz, F.: SVM multiregression for nonlinear channel estimation in multiple-input multiple-output systems. IEEE Trans. Signal Process. 52(8), 2298–2307 (2004)
Zhen, X., Wang, Z., Islam, A., Bhaduri, M., Chan, I., Li, S.: Multi-scale deep networks and regression forests for direct bi-ventricular volume estimation. Med. Image Anal. 30, 120–129 (2016)
Kooi, T., Litjens, G., van Ginneken, B., Gubern-Mrida, A., Snchez, C.I., Mann, R., den Heeten, A., Karssemeijer, N.: Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 35, 303–312 (2017)
Christ, P.F., Elshaer, M.E.A., Ettlinger, F., Tatavarty, S., Bickel, M., Bilic, P., Rempfler, M., Armbruster, M., Hofmann, F., D’Anastasi, M., Sommer, W.H., Ahmadi, S., Menze, B.H.: Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. CoRR abs/1610.02177
Acuña, E., Rodriguez, C.: On detection of outliers and their effect in supervised classification (2004)
Sun, H., Zhen, X., Bailey, C., Rasoulinejad, P., Yin, Y., Li, S.: Direct estimation of spinal cobb angles by structured multi-output regression. In: Niethammer, M., Styner, M., Aylward, S., Zhu, H., Oguz, I., Yap, P.-T., Shen, D. (eds.) IPMI 2017. LNCS, vol. 10265, pp. 529–540. Springer, Cham (2017). doi:10.1007/978-3-319-59050-9_42
Chollet, F., Keras: (2015). https://github.com/fchollet/keras
S.D.S. Group: Radiographic Measurement Manual. Medtronic Sofamor Danek, USA (2008)
Criminisi, A., Shotton, J., Robertson, D., Konukoglu, E.: Regression forests for efficient anatomy detection and localization in CT studies. In: Menze, B., Langs, G., Tu, Z., Criminisi, A. (eds.) MCV 2010. LNCS, vol. 6533, pp. 106–117. Springer, Heidelberg (2011). doi:10.1007/978-3-642-18421-5_11
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Wu, H., Bailey, C., Rasoulinejad, P., Li, S. (2017). Automatic Landmark Estimation for Adolescent Idiopathic Scoliosis Assessment Using BoostNet. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10433. Springer, Cham. https://doi.org/10.1007/978-3-319-66182-7_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-66182-7_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66181-0
Online ISBN: 978-3-319-66182-7
eBook Packages: Computer ScienceComputer Science (R0)