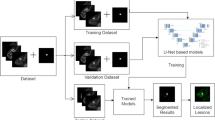
Abstract
Dynamic Contrast Enhanced Magnetic Resonance Imaging (DCE-MRI) is a popular tool for the diagnosis of breast lesions due to its effectiveness, especially in a high risk population. Accurate lesion segmentation is an important step for subsequent analysis, especially for computer aided diagnosis systems. However, manual breast lesion segmentation of (4D) MRI is time consuming, requires experience, and it is prone to interobserver and intraobserver variability. This work proposes a deep learning (DL) framework for segmenting breast lesions in DCE-MRI using a 3D patch based U-Net architecture. We perform different experiments to analyse the effects of class imbalance, different patch sizes, optimizers and loss functions in a cross-validation fashion using 46 images from a subset of a challenging and publicly available dataset not reported to date, that is the TCGA-BRCA. We also compare the proposed U-Net framework with another state-of-the-art approach used for breast lesion segmentation in DCE-MRI, and report better segmentation accuracy with the proposed framework. The results presented in this work have the potential to become a publicly available benchmark for this task.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Badrinarayanan, V., Kendall, A., Cipolla, R.: SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017)
Bria, A., Karssemeijer, N., Tortorella, F.: Learning from unbalanced data: a cascade-based approach for detecting clustered microcalcifications. Med. Image Anal. 18(2), 241–252 (2014)
Chen, M., Zheng, H., Lu, C., Tu, E., Yang, J., Kasabov, N.: A spatio-temporal fully convolutional network for breast lesion segmentation in DCE-MRI. In: Cheng, L., Leung, A.C.S., Ozawa, S. (eds.) ICONIP 2018. LNCS, vol. 11307, pp. 358–368. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-04239-4_32
Clark, K., et al.: The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26(6), 1045–1057 (2013). https://doi.org/10.1007/s10278-013-9622-7
Degani, H., Gusis, V., Weinstein, D., Fields, S., Strano, S.: Mapping pathophysiological features of breast tumors by MRI at high spatial resolution. Nat. Med. 3(7), 780–782 (1997)
DeSantis, C.E., et al.: Breast cancer statistics, 2019. CA Cancer J. Clin. 69(6), 438–451 (2019)
El Adoui, M., Mahmoudi, S., Larhmam, A., Benjelloun, M.: MRI breast tumor segmentation using different encoder and decoder CNN architectures. J. Comput. 8, 52 (2019)
Gubern-Mérida, A., et al.: Automated localization of breast cancer in DCE-MRI. Med. Image Anal. 20(1), 265–274 (2014)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3431–3440 (2015)
Piantadosi, G., Marrone, S., Galli, A., Sansone, M., Sansone, C.: DCE-MRI breast lesions segmentation with a 3TP U-Net deep convolutional neural network. In: 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), pp. 628–633. IEEE (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Subbhuraam, V.S., Ng, E., Acharya, U.R., Faust, O.: Breast imaging: a survey. World J. Clin. Oncol. 2(4), 171–178 (2011)
Vignati, A., et al.: Performance of a fully automatic lesion detection system for breast DCE-MRI. J. Magn. Reson. Imaging JMRI 34(6), 1341–1351 (2011)
Zhang, J., Gao, Y., Park, S.H., Zong, X., Lin, W., Shen, D.: Structured learning for 3-D perivascular space segmentation using vascular features. IEEE Trans. Biomed. Eng. 64(12), 2803–2812 (2017)
Zhang, J., Saha, A., Zhu, Z., Mazurowski, M.A.: Hierarchical convolutional neural networks for segmentation of breast tumors in MRI with application to radiogenomics. IEEE Trans. Med. Imaging 38, 435–447 (2019)
Zhang, L., Luo, Z., Chai, R., Arefan, D., Sumkin, J., Wu, S.: Deep-learning method for tumor segmentation in breast DCE-MRI. In: Chen, P.H., Bak, P.R. (eds.) Medical Imaging 2019: Imaging Informatics for Healthcare, Research, and Applications, vol. 10954, pp. 97–102. International Society for Optics and Photonics, SPIE (2019)
Zheng, Y., Baloch, S., Englander, S., Schnall, M.D., Shen, D.: Segmentation and classification of breast tumor using dynamic contrast-enhanced MR images. In: Ayache, N., Ourselin, S., Maeder, A. (eds.) MICCAI 2007. LNCS, vol. 4792, pp. 393–401. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-75759-7_48
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Acknowledgments
This work was partially supported by the project ICEBERG: Image Computing for Enhancing Breast Cancer Radiomics (RTI2018-096333-B-I00, Spanish Ministry). We would also like to thank the TCGA Breast Phenotype Research Group for providing the computer-extracted lesion segmentation data used in this study, which comes from the University of Chicago lab of Maryellen Giger.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Khaled, R., Vidal, J., Martí, R. (2021). Deep Learning Based Segmentation of Breast Lesions in DCE-MRI. In: Del Bimbo, A., et al. Pattern Recognition. ICPR International Workshops and Challenges. ICPR 2021. Lecture Notes in Computer Science(), vol 12661. Springer, Cham. https://doi.org/10.1007/978-3-030-68763-2_32
Download citation
DOI: https://doi.org/10.1007/978-3-030-68763-2_32
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-68762-5
Online ISBN: 978-3-030-68763-2
eBook Packages: Computer ScienceComputer Science (R0)