ABstract
Background
Distant metastasis is the leading cause of death in patients with colorectal cancer (CRC). Tumor dissemination for metastasis formation occurs in advanced cancers and also during early stages of tumorigenesis. Here, we investigated the genes involved in early metastatic seeding of CRC using gene expression analysis.
Patients and Methods
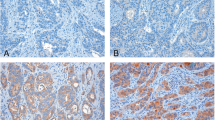
We performed a cDNA microarray using specimens resected from stages I–II CRC with and without metachronous metastatic recurrence. For the candidate genes, we immunohistochemically validated protein expression using a tissue microarray of stages I–III CRC.
Results
The expression of TROP2, VWCE, and BMP7 was upregulated in the recurrence group rather than in the non-recurrence group. Protein expression analysis revealed significant association of these genes with distant metastatic recurrence. The specimens with high expression of BMP7 showed worse recurrence-free survival (RFS; p = 0.02). Those with high expression of TROP2 and VWCE showed worse overall survival (OS) and RFS (TROP2: p = 0.01 and p = 0.03; VWCE: p < 0.05 and p < 0.001, respectively). In the multivariate analysis, high expression of VWCE and BMP7 was an independent predictor of recurrence [VWCE: hazard ratio (HR) 3.41, p < 0.001; BMP7: HR 2.93, p = 0.005]. In contrast, TROP2 was an independent prognostic factor for OS (HR 4.58, p = 0.03).
Conclusions
Gene expression analysis revealed that TROP2, VWCE, and BMP7 were involved in early metastatic seeding. The high expression of these genes may warrant careful surveillance or adjuvant therapy, even in stages I–II CRC cases.




Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502.
DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–71.
Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–17.
Robinson JR, Newcomb PA, Hardikar S, Cohen SA, Phipps AI. Stage IV colorectal cancer primary site and patterns of distant metastasis. Cancer Epidemiol. 2017;48:92–5.
Osterman E, Hammarstrom K, Imam I, Osterlund E, Sjoblom T, Glimelius B. Recurrence risk after radical colorectal cancer surgery-less than before, but how high is it? Cancers (Basel). 2020;12(11):3308.
Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138(6):2059–72.
Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149(5):1177-1190.e3.
Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7.
Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA. 2008;105:4283–8.
Yates LR, Knappskog S, Wedge D, et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32(2):169-184.e7.
Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54.
Sanger N, Effenberger KE, Riethdorf S, et al. Disseminated tumor cells in the bone marrow of patients with ductal carcinoma in situ. Int J Cancer. 2011;129(10):2522–6.
Rhim AD, Mirek ET, Aiello NM, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(12):349–61.
Eyles J, Puaux AL, Wang X, et al. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J Clin Invest. 2010;120(6):2030–9.
Hu Z, Ding J, Ma Z, et al. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet. 2019;51(7):1113–22.
Fumagalli A, Drost J, Suijkerbuijk SJ, et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc Natl Acad Sci USA. 2017;114(12):E2357–64.
Watanabe T, Itabashi M, Shimada Y, et al. Japanese society for cancer of the colon and rectum (JSCCR) guidelines 2014 for treatment of colorectal cancer. Int J Clin Oncol. 2015;20(2):207–39.
Bando H, Okamoto W, Fukui T, Yamanaka T, Akagi K, Yoshino T. Utility of the quasi-monomorphic variation range in unresectable metastatic colorectal cancer patients. Cancer Sci. 2018;109(11):3411–5.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 2009;4(1):44–57.
Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13.
Choueiri TK, Figueroa DJ, Fay AP, et al. Correlation of PD-L1 tumor expression and treatment outcomes in patients with renal cell carcinoma receiving sunitinib or pazopanib: results from COMPARZ, a randomized controlled trial. Clin Cancer Res. 2015;21(5):1071–7.
Antonio C, Tiago CS, Catharina O, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71.
Michael IL, Wolfgang H, Simon A. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550.25.
van der Stok EP, Spaander MCW, Grünhagen DJ, Verhoef C, Kuipers EJ. Surveillance after curative treatment for colorectal cancer. Nat Rev Clin Oncol. 2017;14(5):297–315.
Shifu C, Yanqing Z, Yaru C, Jia G. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90.
Daehwan K, Joseph MP, Chanhee P, Christopher B, Steven LS. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–15.
Yang L, Gordon KS, Wei S. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2017;14(5):297–315.
Ohmachi T, Tanaka F, Mimori K, Inoue H, Yanaga K, Mori M. Clinical significance of TROP2 expression in colorectal cancer. Clin Cancer Res. 2006;12(10):3057–63.
Fang YJ, Lu ZH, Wang GQ, Pan ZZ, Zhou ZW, Yun JP, et al. Elevated expressions of MMP7, TROP2, and survivin are associated with survival, disease recurrence, and liver metastasis of colon cancer. Int J Colorectal Dis. 2009;24(8):875–84.
Wang J, Day R, Dong Y, Weintraub SJ, Michel L. Identification of Trop-2 as an oncogene and an attractive therapeutic target in colon cancers. Mol Cancer Ther. 2008;7(2):280–5.
Zhao P, Zhang Z. TNF-alpha promotes colon cancer cell migration and invasion by upregulating TROP-2. Oncol Lett. 2018;15(3):3820–7.
Cubas R, Zhang S, Li M, Chen C, Yao Q. Trop2 expression contributes to tumor pathogenesis by activating the ERK MAPK pathway. Mol Cancer. 2010;9:253.
Lian Z, Liu J, Li L, Li X, Clayton M, Wu MC, et al. Enhanced cell survival of Hep3B cells by the hepatitis B x antigen effector, URG11, is associated with upregulation of beta-catenin. Hepatology. 2006;43(3):415–24.
Zhang G, Zhu F, Han G, Li Z, Yu Q, Li Z, et al. Silencing of URG11 expression inhibits the proliferation and epithelialmesenchymal transition in benign prostatic hyperplasia cells via the RhoA/ROCK1 pathway. Mol Med Rep. 2018;18(1):391–8.
Pan B, Ye Y, Liu H, Zhen J, Zhou H, Li Y, et al. URG11 regulates prostate cancer cell proliferation, migration, and invasion. Biomed Res Int. 2018;2018:4060728.
Du R, Xia L, Sun S, Lian Z, Zou X, Gao J, et al. URG11 promotes gastric cancer growth and invasion by activation of beta-catenin signalling pathway. J Cell Mol Med. 2010;14(3):621–35.
Peng W, Zhang J, Liu J. URG11 predicts poor prognosis of pancreatic cancer by enhancing epithelial-mesenchymal transition-driven invasion. Med Oncol. 2014;31(7):64.
Liu ZL, Wu J, Wang LX, Yang JF, Xiao GM, Sun HP, et al. Knockdown of upregulated gene 11 (URG11) inhibits proliferation, invasion, and beta-catenin expression in non-small cell lung cancer cells. Oncol Res. 2016;24(3):197–204.
Du R, Huang C, Bi Q, Zhai Y, Xia L, Liu J, et al. URG11 mediates hypoxia-induced epithelial-to-mesenchymal transition by modulation of E-cadherin and beta-catenin. Biochem Biophys Res Commun. 2010;391(1):135–41.
Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19(1):71–88.
Voorneveld PW, Kodach LL, Jacobs RJ, van Noesel CJ, Peppelenbosch MP, Korkmaz KS, et al. The BMP pathway either enhances or inhibits the Wnt pathway depending on the SMAD4 and p53 status in CRC. Br J Cancer. 2015;112(1):122–30.
Motoyama K, Tanaka F, Kosaka Y, Mimori K, Uetake H, Inoue H, et al. Clinical significance of BMP7 in human colorectal cancer. Ann Surg Oncol. 2008;15(5):1530–7.
Cortez MA, Masrorpour F, Ivan C, Zhang J, Younes AI, Lu Y, et al. Bone morphogenetic protein 7 promotes resistance to immunotherapy. Nat Commun. 2020;11(1):4840.
Na YR, Seok SH, Kim DJ, Han JH, Kim TH, Jung H, et al. Bone morphogenetic protein 7 induces mesenchymal-to-epithelial transition in melanoma cells, leading to inhibition of metastasis. Cancer Sci. 2009;100(11):2218–25.
Arvind D, Van KM, Carmen JA, et al. ctDNA applications and integration in colorectal cancer: an NCI colon and rectal-anal task forces whitepaper. Nat Rev Clin Oncol. 2020;17(12):757–70.
Zaman S, Jadid H, Denson AC, Gray JE. Targeting Trop-2 in solid tumors: future prospects. Onco Targets Ther. 2019;12:1781–90.
Bardia A, Mayer IA, Vahdat LT, Tolaney SM, Isakoff SJ, Diamond JR, et al. Sacituzumab Govitecan-hziy in refractory metastatic triple-negative breast cancer. N Engl J Med. 2019;380(8):741–51.
Acknowledgments
We would like to thank N Kobayashi, M. Horigome, A. Nange, and A. Okumura for their technical assistance, and all staff members at the Hokkaido University Hospital. The results shown here are based in part on data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding
This study was supported by JSPS KAKENHI Grant No. JP20K17670.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosures
The authors have no conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sawada, A., Ohira, M., Hatanaka, K.C. et al. Expression Analysis of Early Metastatic Seeding of Colorectal Cancer. Ann Surg Oncol 31, 2101–2113 (2024). https://doi.org/10.1245/s10434-023-14714-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-023-14714-7