Abstract
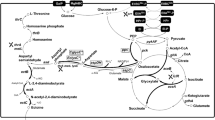
DNA scaffold that enhances the spatial proximity of enzymes and the local concentration of intermediates, is promising tools in optimizing heterologous metabolic pathways for target product biosynthesis. Here, we display the utility of a DNA scaffold system for the production of ectoine in E. coli MWZ003. Three fused enzymes EctA-ZFa, EctB-ZFb, and EctC-ZFc were firstly constructed by fusing enzymes of ectoine synthesis pathway with corresponding zinc finger domains. The copy number of the plasmid-expressing fusions was adapted by substitution of different replicons. Furthermore, a series of modifications were carried out on the DNA scaffold system through optimizing the spacer between enzyme binding sites, the binding direction of fusion enzymes, the repeating unit of DNA scaffolds, the stoichiometric ratio of enzyme binding sites, and the expression level of the rate-limiting enzyme. The optimized DNA scaffold system in the plasmid pFV30 involving use of pMB1 replicon, reverse binding, 11-bp spacer, 4 repeating units, stoichiometric ratio (1:2:2), and enhanced expression of EctB-ZFb increased the ectoine titer and yield, respectively, to 22.79 g/L and 0.65 g/g glucose with increase by 92% compared with that of the control strain. The post-translational strategy based on DNA scaffold was efficient in promoting heterologous synthesis of ectoine, which could also be used in combination with other genetic engineering tools.








Similar content being viewed by others
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code availability
Not applicable.
References
Kogure T, Inui M. Recent advances in metabolic engineering of Corynebacterium glutamicum for bioproduction of value-added aromatic chemicals and natural products. Appl Microbiol Biotechnol. 2018;102(20):8685–705. https://doi.org/10.1007/s00253-018-9289-6.
Cen X, Liu Y, Zhu F, Liu D, Chen Z. Metabolic engineering of Escherichia coli for high production of 1,5-pentanediol via a cadaverine-derived pathway. Metab Eng. 2022;74:168–77. https://doi.org/10.1016/j.ymben.2022.10.012.
Srivastava R, Sahoo L. Cowpea NAC transcription factors positively regulate cellular stress response and balance energy metabolism in yeast via reprogramming of biosynthetic pathways. ACS Synth Biol. 2021;10(9):2286–307. https://doi.org/10.1021/acssynbio.1c00208.
Henke NA, Frohwitter J, Peters-Wendisch P, Wendisch VF. Carotenoid production by recombinant Corynebacterium glutamicum: strain construction, cultivation, extraction, and quantification of carotenoids and terpenes. Methods Mol Biol. 2018;1852:127–41. https://doi.org/10.1007/978-1-4939-8742-9_8.
Xu W, Wang D, Fan J, Zhang L, Ma X, Yao J, Wang Y. Improving squalene production by blocking the competitive branched pathways and expressing rate-limiting enzymes in Rhodopseudomonas palustris. Biotechnol Appl Biochem. 2022;69(4):1502–8. https://doi.org/10.1002/bab.2222.
Chen Y, Wang Z, Chu J, Xi B, Zhuang Y. The glucose RQ-feedback control leading to improved erythromycin production by a recombinant strain Saccharopolyspora erythraea ZL1004 and its scale-up to 372-m3 fermenter. Bioprocess Biosyst Eng. 2015;38(1):105–12. https://doi.org/10.1007/s00449-014-1248-8.
Bomble YJ, Beckham GT, Matthews JF, Nimlos MR, Himmel ME, Crowley MF. Modeling the self-assembly of the cellulosome enzyme complex. J Biol Chem. 2011;286(7):5614–23. https://doi.org/10.1074/jbc.M110.186031.
Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27(8):753–9. https://doi.org/10.1038/nbt.1557.
Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332(6030):680–6. https://doi.org/10.1126/science.1198701.
Peisajovich SG, Garbarino JE, Wei P, Lim WA. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. 2010;328(5976):368–72. https://doi.org/10.1126/science.1182376.
Yaniv O, Shimon LJ, Bayer EA, Lamed R, Frolow F. Scaffoldin-borne family 3b carbohydrate-binding module from the cellulosome of Bacteroides cellulosolvens: structural diversity and significance of calcium for carbohydrate binding. Acta Crystallogr D. 2011;67(Pt 6):506–15. https://doi.org/10.1107/S0907444911011322.
Zeke A, Lukacs M, Lim WA, Remenyi A. Scaffolds: interaction platforms for cellular signalling circuits. Trends Cell Biol. 2009;19(8):364–74. https://doi.org/10.1016/j.tcb.2009.05.007.
Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333(6041):470–4. https://doi.org/10.1126/science.1206938.
Conrado RJ, Wu GC, Boock JT, Xu H, Chen SY, Lebar T, Turnsek J, Tomsic N, Avbelj M, Gaber R, Koprivnjak T, Mori J, Glavnik V, Vovk I, Bencina M, Hodnik V, Anderluh G, Dueber JE, Jerala R, DeLisa MP. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res. 2012;40(4):1879–89. https://doi.org/10.1093/nar/gkr888.
Lee JH, Jung SC, le Bui M, Kang KH, Song JJ, Kim SC. Improved production of L-threonine in Escherichia coli by use of a DNA scaffold system. Appl Environ Microbiol. 2013;79(3):774–82. https://doi.org/10.1128/AEM.02578-12.
Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat Methods. 2007;4(7):571–6. https://doi.org/10.1038/nmeth1058.
Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22(11):1399–408. https://doi.org/10.1038/nbt1029.
Chang HC, Kaiser CM, Hartl FU, Barral JM. De novo folding of GFP fusion proteins: high efficiency in eukaryotes but not in bacteria. J Mol Biol. 2005;353(2):397–409. https://doi.org/10.1016/j.jmb.2005.08.052.
Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388(6640):343–9. https://doi.org/10.1038/41024.
Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. An affinity-based scoring scheme for predicting DNA-binding activities of modularly assembled zinc-finger proteins. Nucleic Acids Res. 2009;37(2):506–15. https://doi.org/10.1093/nar/gkn962.
Negi S, Imanishi M, Matsumoto M, Sugiura Y. New redesigned zinc-finger proteins: design strategy and its application. Chemistry. 2008;14(11):3236–49. https://doi.org/10.1002/chem.200701320.
Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Muller-Lerch F, Fu F, Pearlberg J, Gobel C, Dassie JP, Pruett-Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, McCray PB Jr, Cathomen T, Voytas DF, Joung JK. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31(2):294–301. https://doi.org/10.1016/j.molcel.2008.06.016.
Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods. 2011;8(1):67–9. https://doi.org/10.1038/nmeth.1542.
Becker J, Schafer R, Kohlstedt M, Harder BJ, Borchert NS, Stoveken N, Bremer E, Wittmann C. Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microb Cell Fact. 2013;12:110. https://doi.org/10.1186/1475-2859-12-110.
Galinski EA, Pfeiffer HP, Truper HG. 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid. A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur J Biochem. 1985;149(1):135–9. https://doi.org/10.1111/j.1432-1033.1985.tb08903.x.
Graf R, Anzali S, Buenger J, Pfluecker F, Driller H. The multifunctional role of ectoine as a natural cell protectant. Clin Dermatol. 2008;26(4):326–33.
Kanapathipillai M, Lentzen G, Sierks M, Park CB. Ectoine and hydroxyectoine inhibit aggregation and neurotoxicity of Alzheimer’s beta-amyloid. FEBS Lett. 2005;579(21):4775–80. https://doi.org/10.1016/j.febslet.2005.07.057.
Sydlik U, Gallitz I, Albrecht C, Abel J, Krutmann J, Unfried K. The compatible solute ectoine protects against nanoparticle-induced neutrophilic lung inflammation. Am J Respir Crit Care Med. 2009;180(1):29–35. https://doi.org/10.1164/rccm.200812-1911OC.
Abdel-Aziz H, Wadie W, Abdallah DM, Lentzen G, Khayyal MT. Novel effects of ectoine, a bacteria-derived natural tetrahydropyrimidine, in experimental colitis. Phytomedicine. 2013;20(7):585–91. https://doi.org/10.1016/j.phymed.2013.01.009.
Pastor JM, Salvador M, Argandona M, Bernal V, Reina-Bueno M, Csonka LN, Iborra JL, Vargas C, Nieto JJ, Canovas M. Ectoines in cell stress protection: uses and biotechnological production. Biotechnol Adv. 2010;28(6):782–801. https://doi.org/10.1016/j.biotechadv.2010.06.005.
Sauer T, Galinski EA. Bacterial milking: a novel bioprocess for production of compatible solutes. Biotechnol Bioeng. 1998;59(1):128. https://doi.org/10.1002/(sici)1097-0290(19980705)59:1%3c128::aid-bit17%3e3.0.co;2-e.
Schubert T, Maskow T, Benndorf D, Harms H, Breuer U. Continuous synthesis and excretion of the compatible solute ectoine by a transgenic, nonhalophilic bacterium. Appl Environ Microbiol. 2007;73(10):3343–7. https://doi.org/10.1128/AEM.02482-06.
Perez-Garcia F, Ziert C, Risse JM, Wendisch VF. Improved fermentative production of the compatible solute ectoine by Corynebacterium glutamicum from glucose and alternative carbon sources. J Biotechnol. 2017;258:59–68. https://doi.org/10.1016/j.jbiotec.2017.04.039.
Zhang S, Fang Y, Zhu L, Li H, Wang Z, Li Y, Wang X. Metabolic engineering of Escherichia coli for efficient ectoine production. Syst Microbiol Biomanuf. 2021;004:001. https://doi.org/10.1007/s43393-021-00031-1.
Fang Y, Wang J, Ma W, Yang J, Zhang H, Zhao L, Chen S, Zhang S, Hu X, Li Y, Wang X. Rebalancing microbial carbon distribution for L-threonine maximization using a thermal switch system. Metab Eng. 2020;61:33–46. https://doi.org/10.1016/j.ymben.2020.01.009.
Selas Castiñeiras T, Williams SG, Hitchcock A, Cole JA, Smith DC, Overton TW. Development of a generic β-lactamase screening system for improved signal peptides for periplasmic targeting of recombinant proteins in Escherichia coli. Sci Rep. 2018;8(1):6986. https://doi.org/10.1038/s41598-018-25192-3.
Ooi AT, Stains CI, Ghosh I, Segal DJ. Sequence-enabled reassembly of beta-lactamase (SEER-LAC): a sensitive method for the detection of double-stranded DNA. Biochemistry. 2006;45(11):3620–5. https://doi.org/10.1021/bi0517032.
Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252(5007):809–17. https://doi.org/10.1126/science.2028256.
Stains CI, Porter JR, Ooi AT, Segal DJ, Ghosh I. DNA sequence-enabled reassembly of the green fluorescent protein. J Am Chem Soc. 2005;127(31):10782–3. https://doi.org/10.1021/ja051969w.
Funding
This work is supported by the National Key Research and Development Program of China (2018YFA0900300).
Author information
Authors and Affiliations
Contributions
ZL and XW conceived and designed the research. ZL, YF, HL, SZ, YL, DH and YW conducted experiments. ZL and XW analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Z., Fang, Y., Li, H. et al. DNA scaffold assisted ectoine production in Escherichia coli. Syst Microbiol and Biomanuf 4, 188–202 (2024). https://doi.org/10.1007/s43393-023-00180-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-023-00180-5