Abstract
Microbes thrive and, in turn, influence the earth’s environment, but most are poorly understood because of our limited capacity to reveal their natural diversity and function. Developing novel tools and effective strategies are critical to ease this dilemma and will help to understand their roles in ecology and human health. Recently, droplet microfluidics is emerging as a promising technology for microbial studies with value in microbial cultivating, screening, and sequencing. This review aims to provide an overview of droplet microfluidics techniques for microbial research. First, some critical points or steps in the microfluidic system are introduced, such as droplet stabilization, manipulation, and detection. We then highlight the recent progress of droplet-based methods for microbiological applications, from high-throughput single-cell cultivation, screening to the targeted or whole-genome sequencing of single cells.




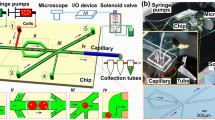
(Adapted from Park et al. 2011. PLOS ONE 6: e17019. followed the terms of the Creative Commons Attribution License). c Schematic of using a co-cultivation strategy for bear oral microbiome screening. d The microfluidic droplets with the encapsulation of Staphylococcus aureus (S. aureus) and oral microbes after cultivation (scale bar 50 μm) (Fig. 4C and 4D reprinted from Terekhov et al. 2018. Proc Natl Acad Sci USA 115:9551–9556. followed Creative Commons Attribution-NonCommercial-NoDerivatives License 4.0 (CC BY-NC-ND))






Similar content being viewed by others
References
Abate AR, Weitz DA (2011) Faster multiple emulsification with drop splitting. Lab Chip 11:1911–1915
Abate AR, Hung T, Mary P, Agresti JJ, Weitz DA (2010) High-throughput injection with microfluidics using picoinjectors. Proc Natl Acad Sci USA 107:19163–19166
Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD, Weitz DA (2010) Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc Natl Acad Sci USA 107:4004–4009
Autour A, Ryckelynck M (2017) Ultrahigh-throughput improvement and discovery of enzymes using droplet-based microfluidic screening. Micromachines 8:128
Bai Y, He X, Liu D, Patil SN, Bratton D, Huebner A, Hollfelder F, Abell C, Huck WTS (2010) A double droplet trap system for studying mass transport across a droplet-droplet interface. Lab Chip 10:1281–1285
Baret J (2012) Surfactants in droplet-based microfluidics. Lab Chip 12:422–433
Baret J, Miller OJ, Taly V, Ryckelynck M, El-Harrak A, Frenz L, Rick C, Samuels ML, Hutchison JB, Agresti JJ, Link DR, Weitz DA, Griffiths AD (2009) Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip 9:1850–1858
Blainey PC (2013) The future is now: single-cell genomics of bacteria and archaea. FEMS Microbiol Rev 37:407–427
Blattman SB, Jiang W, Oikonomou P, Tavazoie S (2020) Prokaryotic single-cell RNA sequencing by in situ combinatorial indexing. Nat Microbiol 5:1–10
Boedicker JQ, Li L, Kline TR, Ismagilov RF (2008) Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip 8:1265–1272
Boedicker JQ, Vincent ME, Ismagilov RF (2009) Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew Chem Int Edit 48:5908–5911
Boers SA, Hays JP, Jansen R (2015) Micelle PCR reduces chimera formation in 16S rRNA profiling of complex microbial DNA mixtures. Sci Rep-UK 5:14181
Burmeister A, Grünberger A (2020) Microfluidic cultivation and analysis tools for interaction studies of microbial co-cultures. Curr Opin Biotech 62:106–115
Burmeister A, Hilgers F, Langner A, Westerwalbesloh C, Kerkhoff Y, Tenhaef N, Drepper T, Kohlheyer D, von Lieres E, Noack S, Grünberger A (2019) A microfluidic co-cultivation platform to investigate microbial interactions at defined microenvironments. Lab Chip 19:98–110
Chen D, Liu S, Du W (2019) Chemotactic screening of imidazolinone-degrading bacteria by microfluidic SlipChip. J Hazard Mater 366:512–519
Chijiiwa R, Hosokawa M, Kogawa M, Nishikawa Y, Ide K, Sakanashi C, Takahashi K, Takeyama H (2020) Single-cell genomics of uncultured bacteria reveals dietary fiber responders in the mouse gut microbiota. Microbiome 8:5
Chiou PY, Ohta AT, Wu MC (2005) Massively parallel manipulation of single cells and microparticles using optical images. Nature 436:370–372
Chiu FWY, Stavrakis S (2019) High-throughput droplet-based microfluidics for directed evolution of enzymes. Electrophoresis 40:2860–2872
Chowdhury MS, Zheng W, Kumari S, Heyman J, Zhang X, Dey P, Weitz DA, Haag R (2019) Dendronized fluorosurfactant for highly stable water-in-fluorinated oil emulsions with minimal inter-droplet transfer of small molecules. Nat Commun 10:4546
Christopher GF, Anna SL (2007) Microfluidic methods for generating continuous droplet streams. J Phys D Appl Phys 40:R319–R336
Clausell-Tormos J, Lieber D, Baret JC, El-Harrak A, Miller OJ, Frenz L, Blouwolff J, Humphry KJ, Koster S, Duan H, Holtze C, Weitz DA, Griffiths AD, Merten CA (2008) Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem Biol 15:427–437
Courtois F, Olguin LF, Whyte G, Theberge AB, Huck WTS, Hollfelder F, Abell C (2009) Controlling the retention of small molecules in emulsion microdroplets for use in cell-based bssays. Anal Chem 81:3008–3016
Cramer C, Fischer P, Windhab EJ (2004) Drop formation in a co-flowing ambient fluid. Chem Eng Sci 59:3045–3058
Daniel R (2005) The metagenomics of soil. Nat Rev Microbiol 3:470–478
Davis KER, Sangwan P, Janssen PH (2011) Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming soil bacteria. Environ Microbiol 13:798–805
De Bourcy CFA, De Vlaminck I, Kanbar JN, Wang J, Gawad C, Quake SR (2014) A quantitative comparison of single-cell whole genome amplification methods. PLoS ONE 9:e105585
Eastburn DJ, Huang Y, Pellegrino M, Sciambi A, Ptáček LJ, Abate AR (2015) Microfluidic droplet enrichment for targeted sequencing. Nucleic Acids Res 43:e86
Falkowski PG, Barber RT, Smetacek V (1998) Biogeochemical controls and feedbacks on ocean primary production. Science 281:200–207
Fenneteau J, Chauvin D, Griffiths AD, Nizak C, Cossy J (2017) Synthesis of new hydrophilic rhodamine based enzymatic substrates compatible with droplet-based microfluidic assays. Chem Commun 53:5437–5440
Frenz L, Blank K, Brouzes E, Griffiths AD (2009) Reliable microfluidic on-chip incubation of droplets in delay-lines. Lab Chip 9:1344–1348
Fu Y, Li C, Lu S, Zhou W, Tang F, Xie XS, Huang Y (2015) Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification. Proc Natl Acad Sci USA 112:11923–11928
Garcia SL (2016) Mixed cultures as model communities: hunting for ubiquitous microorganisms, their partners, and interactions. Aquat Microb Ecol 77:79–85
Gielen F, Hours R, Emond S, Fischlechner M, Schell U, Hollfelder F (2016) Ultrahigh-throughput–directed enzyme evolution by absorbance-activated droplet sorting (AADS). Proc Natl Acad Sci USA 113:E7383–E7389
Girault M, Kim H, Arakawa H, Matsuura K, Odaka M, Hattori A, Terazono H, Yasuda K (2017) An on-chip imaging droplet-sorting system: a real-time shape recognition method to screen target cells in droplets with single cell resolution. Sci Rep-UK 7:40072
Griffiths AD, Tawfik DS (2006) Miniaturising the laboratory in emulsion droplets. Trends Biotechnol 24:395–402
Gruner P, Riechers B, Chacòn Orellana LA, Brosseau Q, Maes F, Beneyton T, Pekin D, Baret J (2015) Stabilisers for water-in-fluorinated-oil dispersions: Key properties for microfluidic applications. Curr Opin Colloid In 20:183–191
Gruner P, Riechers B, Semin B, Lim J, Johnston A, Short K, Baret J (2016) Controlling molecular transport in minimal emulsions. Nat Commun 7:10392
Guo MT, Rotem A, Heyman JA, Weitz DA (2012) Droplet microfluidics for high-throughput biological assays. Lab Chip 12:2146–2155
Habbu P, Warad V, Shastri R, Madagundi S, Kulkarni VH (2016) Antimicrobial metabolites from marine microorganisms. Chin J Nat Medicines 14:101–116
Hassan S, Zhang X (2020) Microfluidics as an emerging platform for tackling antimicrobial resistance (AMR): a Review. Curr Anal Chem 16:41–51
He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu S, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, Nelson KE, Lux R, Shi W (2015) Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA 112:244–249
Holland-Moritz DA, Wismer MK, Mann BF, Farasat I, Devine P, Guetschow ED, Mangion I, Welch CJ, Moore JC, Sun S, Kennedy RT (2020) Mass activated droplet sorting (MADS) enables high-throughput screening of enzymatic reactions at nanoliter scale. Angew Chem Int Edit 59:4470–4477
Holtze C, Rowat AC, Agresti JJ, Hutchison JB, Angilè FE, Schmitz CHJ, Köster S, Duan H, Humphry KJ, Scanga RA, Johnson JS, Pisignano D, Weitz DA (2008) Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip 8:1632–1639
Hori M, Fukano H, Suzuki Y (2007) Uniform amplification of multiple DNAs by emulsion PCR. Biochem Bioph Res Co 352:323–328
Hosokawa M, Nishikawa Y, Kogawa M, Takeyama H (2017) Massively parallel whole genome amplification for single-cell sequencing using droplet microfluidics. Sci Rep-UK 7:5199
Hu B, Xu B, Yun J, Wang J, Xie B, Li C, Yu Y, Lan Y, Zhu Y, Dai X, Huang Y, Huang L, Pan J, Du W (2020) High-throughput single-cell cultivation reveals the underexplored rare biosphere in deep-sea sediments along the Southwest Indian Ridge. Lab Chip 20:363–372
Hultman J, Tamminen M, Pärnänen K, Cairns J, Karkman A, Virta M (2018) Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol Ecol 94: fiy038
Huys GR, Raes J (2018) Go with the flow or solitary confinement: a look inside the single-cell toolbox for isolation of rare and uncultured microbes. Curr Opin Microbiol 44:1–8
Jakiela S, Kaminski TS, Cybulski O, Weibel DB, Garstecki P (2013) Bacterial growth and adaptation in microdroplet chemostats. Angew Chem Int Edit 52:8908–8911
Jarosz DF, Brown JCS, Walker GA, Datta MS, Ung WL, Lancaster AK, Rotem A, Chang A, Newby GA, Weitz DA, Bisson LF, Lindquist S (2014) Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell 158:1083–1093
Ji P, Zhang Y, Wang J, Zhao F (2017) MetaSort untangles metagenome assembly by reducing microbial community complexity. Nat Commun 8:14306
Jian X, Guo X, Wang J, Tan ZL, Xing X, Wang L, Zhang C (2020) Microbial microdroplet culture system (MMC): an integrated platform for automated, high-throughput microbial cultivation and adaptive evolution. Biotechnol Bioeng 117:1724–1737
Jiang C, Dong L, Zhao J, Hu X, Shen C, Qiao Y, Zhang X, Wang Y, Ismagilov RF, Liu S, Du W (2016) High-throughput single-cell cultivation on microfluidic streak plates. Appl Environ Microb 82:2210–2218
Kaeberlein T, Lewis K, Epstein SS (2002) Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127–1129
Kaminski TS, Scheler O, Garstecki P (2016) Droplet microfluidics for microbiology: techniques, applications and challenges. Lab Chip 16:2168–2187
Kaushik AM, Hsieh K, Chen L, Shin DJ, Liao JC, Wang T (2017) Accelerating bacterial growth detection and antimicrobial susceptibility assessment in integrated picoliter droplet platform. Biosens Bioelectron 97:260–266
Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF (2008) Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA 105:18188–18193
Kintses B, Hein C, Mohamed MF, Fischlechner M, Courtois F, Lainé C, Hollfelder F (2012) Picoliter cell lysate assays in microfluidic droplet compartments for directed enzyme evolution. Chem Biol 19:1001–1009
Köster S, Angilè FE, Duan H, Agresti JJ, Wintner A, Schmitz C, Rowat AC, Merten CA, Pisignano D, Griffiths AD, Weitz DA (2008) Drop-based microfluidic devices for encapsulation of single cells. Lab Chip 8:1110–1115
Lagier J, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, Dubourg G, Durand G, Mourembou G, Guilhot E, Togo A, Bellali S, Bachar D, Cassir N, Bittar F, Delerce J et al (2016) Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 1:16203
Lan F, Demaree B, Ahmed N, Abate AR (2017) Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat Biotechnol 35:640–646
Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S et al (2013) Antibiotic resistance-the need for global solutions. Lancet Infect Dis 13:1057–1098
Lewis K (2013) Platforms for antibiotic discovery. Nat Rev Drug Discov 12:371–387
Liao S, Tao X, Ju Y, Feng J, Du W, Wang Y (2017) Multichannel dynamic interfacial printing: an alternative multicomponent droplet generation technique for lab in a drop. Acs Appl Mater Inter 9:43545–43552
Link DR, Anna SL, Weitz DA, Stone HA (2004) Geometrically mediated breakup of drops in microfluidic devices. Phys Rev Lett 92:054503
Liu W, Kim HJ, Lucchetta EM, Du W, Ismagilov RF (2009) Isolation, incubation, and parallel functional testing and identification by FISH of rare microbial single-copy cells from multi-species mixtures using the combination of chemistrode and stochastic confinement. Lab Chip 9:2153–2162
Lok C (2015) Mining the microbial dark matter. Nature 522:270–273
Luka G, Ahmadi A, Najjaran H, Alocilja E, DeRosa M, Wolthers K, Malki A, Aziz H, Althani A, Hoorfar M (2015) Microfluidics integrated biosensors: a leading technology towards lab-on-a-chip and sensing applications. Sensors 15:30011–30031
Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL (2013) Practical innovations for high-throughput amplicon sequencing. Nat Methods 10:999–1002
Ma L, Kim J, Hatzenpichler R, Karymov MA, Hubert N, Hanan IM, Chang EB, Ismagilov RF (2014) Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in Human Microbiome Project’s Most Wanted taxa. Proc Natl Acad Sci USA 111:9768–9773
Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ (2010) Target-enrichment strategies for next-generation sequencing. Nat Methods 7:111–118
Marcy Y, Ishoey T, Lasken RS, Stockwell TB, Walenz BP, Halpern AL, Beeson KY, Goldberg SMD, Quake SR (2007) Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet 3:e155
Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, Quake SR (2007) Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA 104:11889–11894
Martin K, Henkel T, Baier V, Grodrian A, Schön T, Roth M, Michael Köhler J, Metze J (2003) Generation of larger numbers of separated microbial populations by cultivation in segmented-flow microdevices. Lab Chip 3:202–207
Mashaghi S, Abbaspourrad A, Weitz DA, van Oijen AM (2016) Droplet microfluidics: a tool for biology, chemistry and nanotechnology. TRAC-Trend Anal Chem 82:118–125
Mayr LM, Bojanic D (2009) Novel trends in high-throughput screening. Curr Opin Pharmacol 9:580–588
Mazutis L, Baret J, Treacy P, Skhiri Y, Araghi AF, Ryckelynck M, Taly V, Griffiths AD (2009) Multi-step microfluidic droplet processing: kinetic analysis of an in vitro translated enzyme. Lab Chip 9:2902–2908
Nai C, Meyer V (2018) From axenic to mixed cultures: technological advances accelerating a paradigm shift in microbiology. Trends Microbiol 26:538–554
Najah M, Calbrix R, Mahendra-Wijaya IP, Beneyton T, Griffiths AD, Drevelle A (2014) Droplet-based microfluidics platform for ultra-high-throughput bioprospecting of cellulolytic microorganisms. Chem Biol 21:1722–1732
Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, Belanger A, Kanigan T, Lewis K, Epstein SS (2010) Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microb 76:2445–2450
Obexer R, Godina A, Garrabou X, Mittl PRE, Baker D, Griffiths AD, Hilvert D (2017) Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat Chem 9:50–56
Ostafe R, Prodanovic R, Lloyd Ung W, Weitz DA, Fischer R (2014) A high-throughput cellulase screening system based on droplet microfluidics. Biomicrofluidics 8:41102
Pan M, Rosenfeld L, Kim M, Xu M, Lin E, Derda R, Tang SKY (2014) Fluorinated pickering emulsions impede interfacial transport and form rigid interface for the growth of anchorage-dependent cells. Acs Appl Mater Inter 6:21446–21453
Park J, Kerner A, Burns MA, Lin XN (2011) Microdroplet-enabled highly parallel co-cultivation of microbial communities. PLoS ONE 6:e17019
Pham VHT, Kim J (2012) Cultivation of unculturable soil bacteria. Trends Biotechnol 30:475–484
Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR (2012) Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 84:1003–1011
Prakadan SM, Shalek AK, Weitz DA (2017) Scaling by shrinking: empowering single-cell “omics” with microfluidic devices. Nat Rev Genet 18:345–361
Pratt SL, Zath GK, Akiyama T, Williamson KS, Franklin MJ, Chang CB (2019) DropSOAC: stabilizing microfluidic drops for time-lapse quantification of single-cell bacterial physiology. Front Microbiol 10:2112
Qiao Y, Zhao X, Zhu J, Tu R, Dong L, Wang L, Dong Z, Wang Q, Du W (2018) Fluorescence-activated droplet sorting of lipolytic microorganisms using a compact optical system. Lab Chip 18:190–196
Qin H, Wang S, Feng K, He Z, Virta MPJ, Hou W, Dong H, Deng Y (2019) Unraveling the diversity of sedimentary sulfate-reducing prokaryotes (SRP) across Tibetan saline lakes using epicPCR. Microbiome 7:71
Rausch P, Rühlemann M, Hermes BM, Doms S, Dagan T, Dierking K, Domin H, Fraune S, von Frieling J, Hentschel U, Heinsen F, Höppner M, Jahn MT, Jaspers C, Kissoyan KAB, Langfeldt D, Rehman A, Reusch TBH, Roeder T, Schmitz RA et al (2019) Comparative analysis of amplicon and metagenomic sequencing methods reveals key features in the evolution of animal metaorganisms. Microbiome 7:133
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM et al (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437
Rinke C, Lee J, Nath N, Goudeau D, Thompson B, Poulton N, Dmitrieff E, Malmstrom R, Stepanauskas R, Woyke T (2014) Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat Protoc 9:1038–1048
Roach LS, Song H, Ismagilov RF (2005) Controlling nonspecific protein adsorption in a plug-based microfluidic system by controlling interfacial chemistry using fluorous-phase surfactants. Anal Chem 77:785–796
Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, Coenen S, Cohen J, Findlay D, Gyssens I, Heure OE, Kahlmeter G, Kruse H, Laxminarayan R, Liébana E, López-Cerero L, MacGowan A, Martins M, Rodríguez-Baño J, Rolain JM, Segovia C et al (2015) The global threat of antimicrobial resistance: science for intervention. New Microbes and New Infections 6:22–29
Saleski TE, Kerner AR, Chung MT, Jackman CM, Khasbaatar A, Kurabayashi K, Lin XN (2019) Syntrophic co-culture amplification of production phenotype for high-throughput screening of microbial strain libraries. Metab Eng 54:232–243
Sánchez BJ, Lee J, Kang D (2019) Recent advances in droplet-based microfluidic technologies for biochemistry and molecular biology. Micromachines 10:412
Sciambi A, Abate AR (2015) Accurate microfluidic sorting of droplets at 30 kHz. Lab Chip 15:47–51
Shang L, Cheng Y, Zhao Y (2017) Emerging droplet microfluidics. Chem Rev 117:7964–8040
Shao K, Ding W, Wang F, Li H, Ma D, Wang H (2011) Emulsion PCR: a high efficient way of PCR amplification of random DNA libraries in aptamer selection. PLoS One 6:e24910
Sharon I, Banfield JF (2013) Genomes from metagenomics. Science 342:1057
Shemesh J, Ben Arye T, Avesar J, Kang JH, Fine A, Super M, Meller A, Ingber DE, Levenberg S (2014) Stationary nanoliter droplet array with a substrate of choice for single adherent/nonadherent cell incubation and analysis. Proc Natl Acad Sci USA 111:11293–11298
Shen C, Xu P, Huang Z, Cai D, Liu S, Du W (2014) Bacterial chemotaxis on SlipChip. Lab Chip 14:3074–3080
Singer E, Bushnell B, Coleman-Derr D, Bowman B, Bowers RM, Levy A, Gies EA, Cheng J, Copeland A, Klenk H, Hallam SJ, Hugenholtz P, Tringe SG, Woyke T (2016) High-resolution phylogenetic microbial community profiling. ISME J 10:2020–2032
Skhiri Y, Gruner P, Semin B, Brosseau Q, Pekin D, Mazutis L, Goust V, Kleinschmidt F, El Harrak A, Hutchison JB, Mayot E, Bartolo J, Griffiths AD, Taly V, Baret J (2012) Dynamics of molecular transport by surfactants in emulsions. Soft Matter 8:10618–10627
Song H, Chen DL, Ismagilov RF (2006) Reactions in droplets in microfluidic channels. Angew Chem Int Edit 45:7336–7356
Spencer SJ, Tamminen MV, Preheim SP, Guo MT, Briggs AW, Brito IL, Weitz DA, Pitkänen LK, Vigneault F, Virta MP, Alm EJ, (2016) Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J 10:427–436
Tanaka T, Kawasaki K, Daimon S, Kitagawa W, Yamamoto K, Tamaki H, Tanaka M, Nakatsu CH, Kamagata Y (2014) A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl Environ Microb 80:7659–7666
Tao Y, Rotem A, Zhang H, Chang CB, Basu A, Kolawole AO, Koehler SA, Ren Y, Lin JS, Pipas JM, Feldman AB, Wobus CE, Weitz DA (2015) Rapid, targeted and culture-free viral infectivity assay in drop-based microfluidics. Lab Chip 15:3934–3940
Teh S, Lin R, Hung L, Lee AP (2008) Droplet microfluidics. Lab Chip 8:198–220
Terekhov SS, Smirnov IV, Malakhova MV, Samoilov AE, Manolov AI, Nazarov AS, Danilov DV, Dubiley SA, Osterman IA, Rubtsova MP, Kostryukova ES, Ziganshin RH, Kornienko MA, Vanyushkina AA, Bukato ON, Ilina EN, Vlasov VV, Severinov KV, Gabibov AG, Altman S (2018) Ultrahigh-throughput functional profiling of microbiota communities. Proc Natl Acad Sci USA 115:9551–9556
Thorsen T, Roberts RW, Arnold FH, Quake SR (2001) Dynamic pattern formation in a vesicle-generating microfluidic device. Phys Rev Lett 86:4163–4166
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H et al (2004) Environmental genome shotgun sequencing of the sargasso sea. Science 304:66
Wagner O, Thiele J, Weinhart M, Mazutis L, Weitz DA, Huck WTS, Haag R (2016) Biocompatible fluorinated polyglycerols for droplet microfluidics as an alternative to PEG-based copolymer surfactants. Lab Chip 16:65–69
Wang X, Ren L, Su Y, Ji Y, Liu Y, Li C, Li X, Zhang Y, Wang W, Hu Q, Han D, Xu J, Ma B (2017) Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells. Anal Chem 89:12569–12577
Whitesides GM (2006) The origins and the future of microfluidics. Nature 442:368–373
Woyke T, Doud DFR, Schulz F (2017) The trajectory of microbial single-cell sequencing. Nat Methods 14:1045–1054
Xi H, Zheng H, Guo W, Gañán-Calvo AM, Ai Y, Tsao C, Zhou J, Li W, Huang Y, Nguyen N, Tan SH (2017) Active droplet sorting in microfluidics: a review. Lab Chip 17:751–771
Xiao H, Bao Z, Zhao H (2015) High throughput screening and selection methods for directed enzyme evolution. Ind Eng Chem Res 54:4011–4020
Xu Y, Zhao F (2018) Single-cell metagenomics: challenges and applications. Protein Cell 9:501–510
Xu P, Modavi C, Demaree B, Twigg F, Liang B, Sun C, Zhang W, Abate AR (2020) Microfluidic automated plasmid library enrichment for biosynthetic gene cluster discovery. Nucleic Acids Res 48:e48
Yun J, Zheng X, Xu P, Zheng X, Xu J, Cao C, Fu Y, Xu B, Dai X, Wang Y, Liu H, Yi Q, Zhu Y, Wang J, Wang L, Dong Z, Huang L, Huang Y, Du W (2019) Interfacial nanoinjection-based nanoliter single-cell analysis. Small 16:1903739
Zengler K, Toledo G, Rappe M, Elkins J, Mathur EJ, Short JM, Keller M (2002) Nonlinear partial differential equations and applications: Cultivating the uncultured. Proc Natl Acad Sci USA 99:15681–15686
Zhang C, Kim S (2010) Research and application of marine microbial enzymes: status and prospects. Mar Drugs 8:1920–1934
Zhang H, Wang X (2016) Modular co-culture engineering, a new approach for metabolic engineering. Metab Eng 37:114–121
Zhang K, Qin S, Wu S, Liang Y, Li J (2020) Microfluidic systems for rapid antibiotic susceptibility tests (ASTs) at the single-cell level. Chem Sci 11:6352–6361
Zhou N, Sun YT, Chen DW, Du W, Yang H, Liu SJ (2018) Harnessing microfluidic streak plate technique to investigate the gut microbiome of Reticulitermes chinensis. Microbiologyopen 8:e654
Zhu Y, Fang Q (2013) Analytical detection techniques for droplet microfluidics—a review. Anal Chim Acta 787:24–35
Zhu P, Wang L (2017) Passive and active droplet generation with microfluidics: a review. Lab Chip 17:34–75
Acknowledgements
This work was financially supported by National Key Research and Development Program of China (2018YFC0310703), China Ocean Mineral Resources R&D Association (DY135-B-02), National Natural Science Foundation of China (91951103, 91951105, 21822408, 31970091), Key Program of Frontier Sciences of the Chinese Academy of Sciences (QYZDB-SSW-SMC008), and Center for Ocean Mega-Science, Chinese Academy of Sciences (KEXUE2019GZ05).
Author information
Authors and Affiliations
Contributions
WD, LH and XD provided the idea of this review; BH, PX and WD wrote this manuscript; WD, LM, LH, XD, and DC revised this manuscript. BH, WD and PX drew and collected figures and tables. The final manuscript was approved by all of the authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare there is no conflict of interest between them.
Animal and human rights statement
This article does not contain any studies with human participants or animals performed by the authors.
Additional information
SPECIAL TOPIC: Cultivation of uncultured microorganisms.
Edited by Chengchao Chen.
Rights and permissions
About this article
Cite this article
Hu, B., Xu, P., Ma, L. et al. One cell at a time: droplet-based microbial cultivation, screening and sequencing. Mar Life Sci Technol 3, 169–188 (2021). https://doi.org/10.1007/s42995-020-00082-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42995-020-00082-8