Abstract
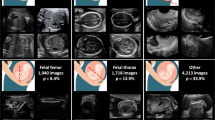
Antenatal (prenatal) care stipulates periodic monitoring of the foetus in alleviating risk factors and improving pregnancy outcomes. Foetal images generated from the ultrasound during prenatal care can be classified into different planes, such as the brain, abdomen, femur, etc., and can be further used for the abnormality detection, disease diagnosis, and foetal growth estimation of the foetus. Therefore, recognizing the foetal plane is one of the initial steps to automate antenatal care. This paper proposes an Ensemble Convolution Neural Network (ECNN) model that combines the base models to classify the foetal planes by using an open-access database containing 12,400 images with six foetal planes. Prior studies on this database involved state-of-the-art CNN methods, and Densenet-169 pre-trained model gave an accuracy of 93.6%. Three pre-trained CNN models (base learners) are trained using the transfer learning approach in the proposed method. The predicted features derived from the base learners are then used to train a Deep Neural Network (DNN) based meta-learner to achieve high classification rates. The stacked ensemble model resulted in an accuracy of \(96\%\), which is better than the accuracy of the individual pre-trained models.







Similar content being viewed by others
Data availability
The data used in the study was obtained from a open access website: https://zenodo.org/record/3904280.
References
Ekabua J, Ekabua K, Njoku C (2011) Proposed framework for making focused antenatal care services accessible: a review of the Nigerian setting. Int Sch Res Not 2011:253964
He F, Wang Y, Xiu Y, Zhang Y, Chen L (2021) Artificial intelligence in prenatal ultrasound diagnosis. Front Med. https://doi.org/10.3389/fmed.2021.729978
Harikumar S, Akhil AS, Kaimal R (2019) A depth-based nearest neighbor algorithm for high-dimensional data classification. Turk. J. Electr. Eng. Comput. Sci. 27(6):4082–4101
Itoo F, Meenakshi Singh S (2021) Comparison and analysis of logistic regression, Naïve Bayes and KNN machine learning algorithms for credit card fraud detection. Int J Inf Technol (Singap) 13(4):1503–1511
Harikumar S (2020) Blended models for nearest neighbour algorithms for high dimensional smart medical data. Smart medical data sensing and IoT systems design in healthcare. IGI Global, Beijing, pp 48–75
Hari Prakash S, Adithya Narayan K, Nair GS, Harikumar S (2022) Perceiving machine learning algorithms to analyze COVID-19 radiographs. In: Proceedings of international conference on recent trends in computing. Springer, pp. 293–305
Bridget ON, Prasad R, Onime C, Ali AA (2021) Drug resistant tuberculosis classification using logistic regression. International Journal of Information Technology (Singapore) 13(2):741–749
Burgos-Artizzu XP, Coronado-Gutiérrez D, Valenzuela-Alcaraz B, Bonet-Carne E, Eixarch E, Crispi F, Gratacós E (2020) Evaluation of deep convolutional neural networks for automatic classification of common maternal fetal ultrasound planes. Sci Rep 10(1):1–12
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Carneiro G, Mateus D, Peter L, Bradley A, Tavares JMR, Belagiannis V, Papa JP, Nascimento JC, Loog M, Lu Z et al (2016) Deep learning and data labeling for medical applications: first international workshop, LABELS 2016, and second international workshop, DLMIA 2016, held in conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Proceedings, vol 10008. Springer
Kalaiselvi T, Padmapriya ST, Sriramakrishnan P, Somasundaram K (2020) Deriving tumor detection models using convolutional neural networks from MRI of human brain scans. Int J Inf Technol (Singap) 12(2):403–408
Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C (2018) A survey on deep transfer learning. In: International conference on artificial neural networks. Springer, pp. 270–279
Gopakumar G, Sai Subrahmanyam GR (2019) Deep learning applications to cytopathology: a study on the detection of malaria and on the classification of leukaemia cell-lines. Handbook of deep learning applications. Springer, Cham, pp 219–257
Mamun MA, Kabir MS, Akter M, Uddin MS (2022) Recognition of human skin diseases using inception-V3 with transfer learning. Int J Inf Technol (Singap) 14(6):3145–3154
Kora P, Ooi CP, Faust O, Raghavendra U, Gudigar A, Chan WY, Meenakshi K, Swaraja K, Plawiak P, Acharya UR (2021) Transfer learning techniques for medical image analysis: a review. Biocybern Biomed Eng 42:79–107
Wu L, Cheng J-Z, Li S, Lei B, Wang T, Ni D (2017) FUIQA: fetal ultrasound image quality assessment with deep convolutional networks. IEEE Trans Cybern 47(5):1336–1349
Walker MC, Willner I, Miguel OX, Murphy MS, El-Chaâr D, Moretti F, Dingwall Harvey AL, Rennicks White R, Muldoon KA, Carrington AM et al (2022) Using deep-learning in fetal ultrasound analysis for diagnosis of cystic hygroma in the first trimester. PLoS One 17(6):0269323
Rueda S, Fathima S, Knight CL, Yaqub M, Papageorghiou AT, Rahmatullah B, Foi A, Maggioni M, Pepe A, Tohka J et al (2013) Evaluation and comparison of current fetal ultrasound image segmentation methods for biometric measurements: a grand challenge. IEEE Trans Med Imaging 33(4):797–813
Heuvel TL, Bruijn D, Korte CL, Ginneken BV (2018) Automated measurement of fetal head circumference using 2D ultrasound images. PLoS One 13(8):0200412
Despeisse M, Ford S (2015) The role of additive manufacturing in improving resource efficiency and sustainability. In: IFIP international conference on advances in production management systems, pp 129–136. Springer
Baumgartner CF, Kamnitsas K, Matthew J, Fletcher TP, Smith S, Koch LM, Kainz B, Rueckert D (2017) SonoNet: real-time detection and localisation of fetal standard scan planes in freehand ultrasound. IEEE Trans Med Imaging 36(11):2204–2215
Chen H, Ni D, Qin J, Li S, Yang X, Wang T, Heng PA (2015) Standard plane localization in fetal ultrasound via domain transferred deep neural networks. IEEE J Biomed Health Inf 19(5):1627–1636
Liu X, Annangi P, Gupta M, Yu B, Padfield D, Banerjee J, Krishnan K (2012) Learning-based scan plane identification from fetal head ultrasound images. In: Bosch JG, Doyley MM (eds) Medical imaging 2012: ultrasonic imaging, tomography, and therapy. society of photo-optical instrumentation engineers (SPIE) conference series, vol 8320. p 83200
Qu R, Xu G, Ding C, Jia W, Sun M (2020) Standard plane identification in fetal brain ultrasound scans using a differential convolutional neural network. IEEE Access 8:83821–83830
Pu B, Li K, Li S, Zhu N (2021) Automatic fetal ultrasound standard plane recognition based on deep learning and IIoT. IEEE Trans Ind Inf 17:7771–7780
Sridar P, Kumar A, Quinton AE, Nanan R, Kim J, Krishnakumar R (2019) Decision fusion-based fetal ultrasound image plane classification using convolutional neural networks. Ultrasound Med Biol 45(5):1259–1273
Wang X, Liu Z, Du Y, Diao Y, Liu P, Lv G, Zhang H (2021) Recognition of fetal facial ultrasound standard plane based on texture feature fusion. Comput Math Methods Med 2021:1–12
Sarwar A, Ali M, Manhas J, Sharma V (2020) Diagnosis of diabetes type-ii using hybrid machine learning based ensemble model. Int J Inf Technol (Singap) 12(2):419–428
Ju C, Bibaut A, Laan M (2018) The relative performance of ensemble methods with deep convolutional neural networks for image classification. J Appl Stat 45(15):2800–2818
Aloysius N, Geetha M (2021) An ensembled scale-space model of deep convolutional neural networks for sign language recognition. Advances in artificial intelligence and data engineering. Springer, Singapore, pp 363–375
Zheng Y, Li C, Zhou X, Chen H, Xu H, Li Y, Zhang H, Li X, Sun H, Huang X et al (2022) Application of transfer learning and ensemble learning in image-level classification for breast histopathology. arXiv preprint arXiv:2204.08311
Xue D, Zhou X, Li C, Yao Y, Rahaman MM, Zhang J, Chen H, Zhang J, Qi S, Sun H (2020) An application of transfer learning and ensemble learning techniques for cervical histopathology image classification. IEEE Access 8:104603–104618
Shorfuzzaman M (2022) An explainable stacked ensemble of deep learning models for improved melanoma skin cancer detection. Multimedia Syst 28(4):1309–1323
Wolpert DH (1992) Stacked generalization. Neural Netw 5(2):241–259
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Huang G, Liu Z, Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Thomas, S., Harikumar, S. An ensemble deep learning framework for foetal plane identification. Int. j. inf. tecnol. 16, 1377–1386 (2024). https://doi.org/10.1007/s41870-023-01709-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41870-023-01709-6