Abstract
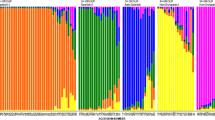
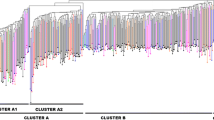
Trifolium L. has around 255 species worldwide. Strawberry clover is one of the most valuable forage crops of the genus Trifolium with two varieties in Iran. In the current study, 40 individuals of eight natural populations of Trifolium fragiferum varieties were examined to reveal genetic diversity and infraspecific relationships with microsatellite markers. Based on five primer sets, a total of 11 alleles were obtained with an average of 2.2 alleles per locus. The polymorphic information content ranged from 0.0 to 0.517 with an average of 0.328. The observed and effective heterozygosity across all loci changed from 0 to 1 and 0 to 0.596, respectively. The gene diversity among the loci varied from 0 to 0.530. Unweighted pair group method with arithmetic mean analyses based on Gower’s distance coefficient grouped accessions into five major clusters but did not separate 40 individuals of T. fragiferum var. fragiferum and T. fragiferum var. pulchellum in distinct groups. Results did not show any clear association between clusters and the geographical origin of varieties. Analysis of molecular variance indicated 84% of the total variation within populations. Genetic differentiation was low in the total population (FST = 0.1), while a high level of gene flow (Nm = 2.247) was estimated among populations. Microsatellite markers evidence suggests that two varieties should not remain separated.



Similar content being viewed by others
References
Abdul-Muneer PM (2014) Application of microsatellite markers in conservation genetics and fisheries management: recent advances in population structure analysis and conservation strategies. Genet Res Int 2014:1–11
Ahsyee RS, Vasiljevic S, Calic I, Zoric M, Karagic D et al (2014) Genetic diversity in red clover (Trifolium pratense L.) using ssr markers. Genetika 46:949–961
Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Wiley, New York
Berzina I, Zhuk A, Veinberga I, Rashal I, Rungis D (2008) Genetic fingerprinting of Latvian red clover (Trifolium pratense L.) cultivars using simple sequence repeat (SSR) markers: comparisons over time and space. Latv J Agron 11:28–32
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Conterato IF, Dall’ Agnol M, Schifino-Wittmann MT, Janke A (2012) Microsatellite diversity and chromosome number in natural populations of Trifolium riograndense Burkart. Sci Agric 69:114–118
Conterato IF, Schifino-Wittmann MT, Guerrad D, Dall’Agnol M (2013) Amphicarpy in Trifolium argentinense: morphological characterization, seed production, reproductive behavior and life strategy. Aust J Bot 61:119–127
Conterato IF, Schifino-Wittmann MT, Guerra D, Büttow MV, Dall’Agnol M et al (2018) Genetic diversity assessed by microsatellite markers in the amphicarpic species Trifolium polymorphum Poir. An Acad Bras Ciênc 90:1685–1693
Dakin EE, Avise JC (2004) Microsatellite null alleles in parentage analysis. Heredity 93:504–509
Dalla Rizza M, Real D, Reyno R, Porro V, Burgueno J et al (2007) Genetic diversity and DNA content of three South American and three Eurasiatic Trifolium species. Genet Mol Biol 30:1118–1124
Davies WE, Young NR (1966) Self-fertility in Trifolium fragiferum. Heredity 21(4):615–624
Dean DA, Wadl PA, Hadziabdic D, Wang X, Triqiano RN (2013) Analyzing microsatellites using the QIAxcel system. Methods Mol Biol 1006:223–243
Dias PMB, Julier B, Sampoux J-P, Barre P, Dall’ Agnol M (2008) Genetic diversity in red clover (Trifolium pratense L.) revealed by morphological and microsatellite (SSR) markers. Euphytica 160:189–205
Dugar YN, Popov VN (2013) Genetic structure and diversity of Ukrainian red clover cultivars revealed by microsatellite markers. Genetics 3:235–242
El-Banna AL, Ghazy MMF (2017) Assessment of genetic components and genetic diversity of six Egyptian clover (Trifolium alexandrinum L.) genotypes using ISSR and URP markers. Egypt J Genet Cytol 46:313–328
Ellison NW, Liston A, Steiner JJ, Williams WM, Taylor NL (2006) Molecular phylogenetics of the clover genus (Trifolium—Leguminosae). Mol Phylogenet Evol 39(3):688–705
Eslami Farouji A, Khodayari H, Assadi M, Özüdogru B, Çetin Ö et al (2018) Numerical taxonomy contributes to delimitation of Iranian and Turkish Hesperis L. (Brassicaceae) species. Phytotaxa 367:101–119
Evans HJ (1962) Chromosome aberrations induced by ionizing radiations. In: Bourne GH, Danielli JF (eds) International review of cytology, vol 13. Academic Press, New York
Gillet JM, Taylor NL (2001) The world of clovers (ed: Collins M). Iowa State University Press, Ames
Gower JC (1971) A general coefficient of similarity and some of its properties. Biometrics 27:857–871. https://doi.org/10.2307/2528823
Grauda D, Avotins K, Fokina O, Kolodinska-Brantestam A, Rashal I (2015) Genetic diversity of white clover (Trifolium repens L.) from the urban area of Riga. Proc Latv Acad Sci Sect Bot 3:132–134
Gupta M, Sharma V, Singh SK, Chahota RK, Sharma TR (2016) Analysis of genetic diversity and structure in a genebank collection of red clover (Trifolium pratense L.) using SSR markers. Plant Genet Resour 15(4):376–379
Haerinasab M, Agahei P (2013) Phenetic analysis of Trifolium resupinatum L. (Persian clover) and Trifolium fragiferum L. (Strawberry clover) using new microsatellite loci. Taxon Biosyst J 5(14):41–51 (in Persian)
Haerinasab M, Rahiminejad MR (2012) A taxonomic revision of the genus Trifolium L. sect. Fragifera Koch (Fabaceae) in Iran. Iran J Bot 18(1):22–30
Haerinasab M, Rahiminejad MR, Ellison NW (2016) Transferability of simple sequence repeat (SSR) markers developed in red clover (Trifolium pratense L.) to some Trifolium species. Iran J Sci Technol Trans A Sci 40(1):59–62
Hammer Ø, Harper D (2006) Paleontological data analysis. Blackwell Publishing, Oxford
Hennink S, Zeven AC (1991) The interpretation of Nei and Shannon-Weaver within population variation indices. Euphytica 51:235–240
Hesamzadeh Hejazi SM, Ziaenasab M (2006) Karyological study on some species of Trifolium genus in Iran. Iran J Bot 39(4):147–151
Issolah R, Abdelguerfi A (1999) Chromosome numbers within some spontaneous populations of 10 Trifolium species in Algeria. Caryologia 52(3–4):151–154
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49(4):725
Kölliker R, Jones ES, Drayton MC, Dupal MP, Forster JW (2001) Development and characterization of simple sequence repeat (SSR) markers for white clover (Trifolium repens L.). Theor Appl Genet 102:416–424
Lefort F, Douglas GC (1999) An efficient micro-method of DNA isolation from mature leaves of four hardwood tree species Acer, Fraxinus, Prunus and Quercus. Ann For Sci 56(3):259–263
Levene H (1949) On a matching problem arising in genetics. Ann Math Stat 20:91–94
Lewontin R (1972) The apportionment of human diversity. Evol Biol 6:381–398
Liu K, Muse S (2004) PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21:2128–2129
Martina S (2007) New microsatellite loci for red clover (Trifolium pratense L.) and study its polymorphism. In: XXVIIth Proceeding of the Eucarpia symposium on improvement of fodder crops and amenity grasses, Copenhagen, pp 217–219
Martins K, Santos JD, Gaiotto FA, Moreno MA, Kageyama PY (2008) Population genetic structure of Copaifera langsdorffii Desf. (Leguminosae–Caesalpinioideae) in forest fragments in “Pontal do Paranapanema”, SP, Brazil. Braz J Bot 31:61–69 (in Portuguese, with abstract in English)
Morley FHW (1963) The mode of pollination in strawberry clover (Trifolium fragiferum). Aust J Exp Agric 3(8):5–8
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 70(12):3321–3323
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Pagnotta MA, Annicchiarico P, Farina A, Proietti S (2011) Characterizing the molecular and morphophysiological diversity of Italian red clover. Euphytica 179(3):393–404
Pathaichindachote W, Panyawut N, Sikaewtung K, Patarapuwadol S, Muangprom A (2019) Genetic diversity and allelic frequency of selected Thai and exotic rice germplasm using SSR markers. Rice Sci 26:393–403
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1(7):215–222
QGIS Development Team (2017) QGIS geographic information system. Open source geospatial foundation project. http://qgis.osgeo.org. Accessed 25 July 2019
Radinovic I, Vasiljevic S, Brankovic G, Ahsyee RS, Momirovic U et al (2017) Molecular characterization of red clover genotypes utilizing microsatellite markers. Chil J Agr Res 77:41–47
Rasouli M, Fattahi Moghaddam MR, Zamani Z, Eimani A, Ebadi A (2014) Evaluation of genetic relationships between almond (Prunus dulcis L) cultivars and genotypes using SSR markers. Iran J Hortic Sci 45(2):151–162
Real D, Dalla Rizza M, Reyno R, Quesenberry KH (2007) Breeding system of the aerial flowers in an amphicarpic clover species: Trifolium polymorphum. Crop Sci 47:1401–1406
Rosso BS, Pagano EM (2005) Evaluation of introduced and naturalized populations of red clover (Trifolium pratense L.) at Pergamino EEA-INTA, Argentina. Genet Resour Crop Evol 52:507–511
Ryman N, Utter F, Hindar K (1995) Introgression, supportive breeding, and genetic conservation. In: Ballou JD, Gilpin M, Foose TJ (eds) Population management for survival and recovery. Columbia University Press, New York, pp 341–365
Salimpour F, Sharifnia F, Mostafavi G, Hajrasoliha SH, Ukhneh E (2008) Chromosome counts and determination of ploidy levels in Iranian species of Trifolium. Chromosome Bot 3:53–63
Santos CAF, Oliveira VR, Rodrigues MA, Ribeiro HLC (2010) Molecular characterization of onion cultivars using microsatellite markers. Pesqui Agropecu Bras 45:49–55 (in Portuguese, with abstract in English)
Sato S, Isobe S, Asamizu E, Ohmido N, Kataoka R et al (2005) Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.). DNA Res 12(5):301–364. https://doi.org/10.1093/dnares/dsi018
Shannon CE, Weaver W (1949) The mathematical theory of communication. University of Illinois Press, Urbana
Sharma R, Kumar B, Arora R, Ahlawat S, Mishra AK et al (2016) Genetic diversity estimates point to immediate efforts for conserving the endangered Tibetan sheep of India. Meta Gene 8:14–20
Shen Z, Zhang K, Ma L, Duan J, Ao Y (2017) Analysis of the genetic relationships and diversity among 11 populations of Xanthoceras sorbifolia using phenotypic and microsatellite marker data. J Biotechnol 26:33–39
Slatkin M (1985) Gene flow in natural populations. Annu Rev Ecol Syst 16:393–430
Slatkin M, Barton NH (1989) A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43(7):1349–1368
Sneath PHA, Sokal RR (1973) Numerical taxonomy. The principles and practice of numerical classification. W.H. Freeman and company press, San Francisco
Stebbins GL Jr (1950) Variation and evolution in plants. Columbia University Press, New York
Taylor N, Gillett J (1988) Crossing and morphological relationships among Trifolium species closely related to Strawberry and Persian clover. Crop Sci 28(4):636–639
Uslu E, Ertuğrul GD, Babac MT (2013) Assessment of genetic diversity in naturally growing 29 Trifolium L. taxa from Bolu Province using RAPD and SSR markers. Turk J Bot 37(4):479–490
Vymyslicky T, Smarda P, Pelikan J, Cholastova T, Nedelnik J et al (2012) Evaluation of the Czech core collection of Trifolium pratense, including morphological, molecular and phytopathological data. Afr J Biotechnol 11:3583–3595
Wright S (1964) The distribution of self-incompatibility alleles in populations. Evolution 18:609–619
Yeh FC, Yang RC, Boyle T (1999) POPGENE, the user-friendly shareware for population genetic analysis. Molecular biology and biotechnology centre, University of Alberta, vol 10, pp 295–301
Yousefi S, Saeidi H, Assadi M (2018) Genetic diversity analysis of red clover (Trifolium pratense L.) in Iran using sequence related amplified polymorphism (SRAP) markers. J Agric Sci Technol 20:373–386
Zhang X, Zhang Y-J, Yan R, Han J-G, Hong F et al (2010) Genetic variation of white clover (Trifolium repens L. collections from China detected by morphological traits, RAPD and SSR. Afr J Biotechnol 9:3032–3041
Zohary M, Heller D (1984) The genus Trifolium. Israel Academy of Sciences and Humanities, Jerusalem
Acknowledgements
The authors of the current research would like to thank Payame Noor University and Shahid Bahonar University of Kerman for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Haerinasab, M., Ali-Farsangi, F., Bordbar, F. et al. Genetic Diversity and Infraspecific Relationships of Trifolium fragiferum L. in Iran. Iran J Sci Technol Trans Sci 44, 345–354 (2020). https://doi.org/10.1007/s40995-020-00834-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40995-020-00834-2