Abstract
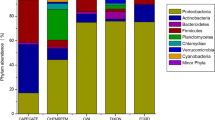
A meta-analysis was carried out using 20 publicly available metagenomic datasets obtained from diverse wastewater samples. A taxonomic assignment was performed, and the diversity measures were calculated. A total of 6,11,942 reads were analysed, and the sequences were assigned to 2605 operational taxonomic units (OTUs) based on ≥ 97% sequence identity. The taxonomic classification carried out at the phyla level indicated that Proteobacteria, Firmicutes, and Bacteroidetes were the dominant bacterial phyla. At the genera level, Comamonas, Pseudomonas, Acidovorax, Arcobacter, and Acinetobacter were prevalent across most wastewaters, forming the core microbial genera. Similarly, Burkholderiaceae, Rhodocyclaceae, Ruminococcaceae, Rhodobacteraceae, and Rhizobiaceae formed the core microbial families across the selected wastewaters. Spearman correlation analysis showed positive and negative correlations between the identified core microbiome, indicating the symbiotic relationship among bacteria involved in bioremediation. Metabolic pathway analysis indicated the presence of genes related to xenobiotics biodegradation and metabolism pathways in the datasets analysed. Majority of the wastewaters harbored an unclassified and uncultured bacterial consortia, indicating a novel microbiome yet to be characterised. Diversity analysis showed similarity in the habitation of microbiome irrespective of the physico-chemical nature and characteristics of wastewaters. As a comprehensive survey of diverse wastewater microbiome, this study provides insights into the core microbial diversity that could improve the existing bioremediation strategies.





Similar content being viewed by others
References
Aggarwal SC, Kumar S (2011) Industrial water demand in India: challenges and implications for water pricing. India Infrastruct Rep 2011:274–281
Andersen SM, Johnsen K, Sørensen J et al (2000) Pseudomonas frederiksbergensis sp. Nov., isolated from soil at a coal gasification site. Int J Syst Evol Microbiol. https://doi.org/10.1099/00207713-50-6-1957
Andrews S (2015) FASTQC a quality control tool for high throughput sequence data. Babraham Inst
Aouidi F, Gannoun H, Ben Othman N et al (2009) Improvement of fermentative decolorization of olive mill wastewater by Lactobacillus paracasei by cheese whey’s addition. Process Biochem 44:597–601. https://doi.org/10.1016/j.procbio.2009.02.014
Ayed L, Hamdi M (2003) Fermentative decolorization of olive mill wastewater by Lactobacillus plantarum. Process Biochem 39:59–65. https://doi.org/10.1016/S0032-9592(02)00314-X
Aziz A, Basheer F, Sengar A et al (2019) Biological wastewater treatment (anaerobic-aerobic) technologies for safe discharge of treated slaughterhouse and meat processing wastewater. Sci Total Environ 686:681–708
Bhattacharya A, Gupta A (2013) Evaluation of Acinetobacter sp. B9 for Cr (VI) resistance and detoxification with potential application in bioremediation of heavy-metals-rich industrial wastewater. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-013-1728-4
Biswas R, Vivekanand V, Saha A et al (2019) Arsenite oxidation by a facultative chemolithotrophic Delftia spp. BAs29 for its potential application in groundwater arsenic bioremediation. Int Biodeterior Biodegrad. https://doi.org/10.1016/j.ibiod.2018.10.006
Boden R, Hutt LP, Rae AW (2017) Reclassification of Thiobacillus aquaesulis (Wood & Kelly 1995) as Annwoodia aquaesulis gen. nov., comb. nov., transfer of Thiobacillus (Beijerinck, 1904) from the Hydrogenophilales to the Nitrosomonadales, proposal of Hydrogenophilalia class. nov. within the ‘Proteobacteria’, and four new families within the orders Nitrosomonadales and Rhodocyclales. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.001927
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bonjoch X, Ballesté E, Blanch AR (2005) Enumeration of bifidobacterial populations with selective media to determine the source of waterborne fecal pollution. Water Res. https://doi.org/10.1016/j.watres.2005.01.018
Buttigieg PL, Ramette A (2014) A guide to statistical analysis in microbial ecology: a community-focused, living review of multivariate data analyses. FEMS Microbiol Ecol. https://doi.org/10.1111/1574-6941.12437
Cappello S, Denaro R, Genovese M et al (2007) Predominant growth of Alcanivorax during experiments on “oil spill bioremediation” in mesocosms. Microbiol Res. https://doi.org/10.1016/j.micres.2006.05.010
Caucci S, Karkman A, Cacace D et al (2016) Seasonality of antibiotic prescriptions for outpatients and resistance genes in sewers and wastewater treatment plant outflow. FEMS Microbiol Ecol 92:1–10. https://doi.org/10.1093/femsec/fiw060
Cea M, Jorquera M, Rubilar O et al (2010) Bioremediation of soil contaminated with pentachlorophenol by Anthracophyllum discolor and its effect on soil microbial community. J Hazard Mater. https://doi.org/10.1016/j.jhazmat.2010.05.013
Chaudhari AU, Paul D, Dhotre D, Kodam KM (2017) Effective biotransformation and detoxification of anthraquinone dye reactive blue 4 by using aerobic bacterial granules. Water Res 122:603–613. https://doi.org/10.1016/j.watres.2017.06.005
Chen TW, Chang SC (2020) Potential microbial indicators for better bioremediation of an aquifer contaminated with vinyl chloride or 1,1-dichloroethene. Water Air Soil Pollut. https://doi.org/10.1007/s11270-020-04538-6
Chen S, Dong X (2004) Acetanaerobacterium elongatum gen. nov., sp. Nov., from paper mill wastewater. Int J Syst Evol Microbiol 54(6):2257–2262
Chen Y, Zhang L, Feng L et al (2019) Exploration of the key functional strains from an azo dye degradation microbial community by DGGE and high-throughput sequencing technology. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-019-05781-z
Chen YL, Wang CH, Yang FC et al (2016) Identification of Comamonas testosteroni as an androgen degrader in sewage. Sci Rep 6:1–13. https://doi.org/10.1038/srep35386
Chikere CB, Okpokwasili GC, Chikere BO (2009) Bacterial diversity in a tropical crude oil-polluted soil undergoing bioremediation. Afr J Biotechnol. https://doi.org/10.4314/ajb.v8i11.60762
Chong J, Liu P, Zhou G, Xia J (2020) Using microbiome analyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. https://doi.org/10.1038/s41596-019-0264-1
Collado L, Levican A, Perez J, Figueras MJ (2011) Arcobacter defluvii sp. nov., isolated from sewage samples. Int J Syst Evol Microbiol 61:2155–2161. https://doi.org/10.1099/ijs.0.025668-0
Cordova-Rosa SM, Dams RI, Cordova-Rosa EV et al (2009) Remediation of phenol-contaminated soil by a bacterial consortium and Acinetobacter calcoaceticus isolated from an industrial wastewater treatment plant. J Hazard Mater 164:61–66. https://doi.org/10.1016/j.jhazmat.2008.07.120
Crini G, Lichtfouse E (2019) Advantages and disadvantages of techniques used for wastewater treatment. Environ Chem Lett 17:145–155. https://doi.org/10.1007/s10311-018-0785-9
Cyprowski M, Stobnicka-Kupiec A, Ławniczek-Wałczyk A et al (2018) Anaerobic bacteria in wastewater treatment plant. Int Arch Occup Environ Health. https://doi.org/10.1007/s00420-018-1307-6
Deblonde T, Cossu-leguille C, Hartemann P (2015) International journal of hygiene and emerging pollutants in wastewater: a review of the literature. Int J Hyg Environ Health 214:442–448. https://doi.org/10.1016/j.ijheh.2011.08.002
Farmer JJ, Arduino MJ, Hickman-Brenner FW (2006) The genera aeromonas and plesiomonas. Prokaryotes 6:564–596
Ferreira AE, Marchetti DP, De Oliveira LM et al (2011) Presence of OXA-23-producing isolates of Acinetobacter baumannii in wastewater from hospitals in Southern Brazil. Microb Drug Resist. https://doi.org/10.1089/mdr.2010.0013
Fisher JC, Levican A, Figueras MJ, McLellan SL (2014) Population dynamics and ecology of Arcobacter in sewage. Front Microbiol 5:1–9. https://doi.org/10.3389/fmicb.2014.00525
Gil-Pulido B, Tarpey E, Almeida EL et al (2018) Evaluation of dairy processing wastewater biotreatment in an IASBR system: aeration rate impacts on performance and microbial ecology. Biotechnol Rep 19:e00263. https://doi.org/10.1016/j.btre.2018.e00263
González G, Herrera G, García MT, Peña M (2001) Biodegradation of phenolic industrial wastewater in a fluidized bed bioreactor with immobilized cells of Pseudomonas putida. Bioresour Technol 80:137–142. https://doi.org/10.1016/S0960-8524(01)00076-1
Goris T, Diekert G (2016) The genus Sulfurospirillum. In: Löffler Frank E, Adrian Lorenz (eds) Organohalide-respiring bacteria. Springer, Cham
Grandclement C, Seyssiecq I, Piram A, et al (2017) From the conventional biological wastewater treatment to hybrid processes, the evaluation of organic micropollutant removal : a review To cite this version : HAL Id : hal-01456484
Harper DA, Ryan PD (2012) PAST: paleontological statistics software package for education and data analysis. Palaeontologia electronica 4:1–9
He LY, He LK, Liu YS et al (2019) Microbial diversity and antibiotic resistome in swine farm environments. Sci Total Environ 685:197–207. https://doi.org/10.1016/j.scitotenv.2019.05.369
Hedges LV, Gurevitch J, Curtis PS (1999) The meta-analysis of response ratios in experimental ecology. Ecology. https://doi.org/10.1890/0012-9658(1999)080
Heylen K, Lebbe L, de Vos P (2008) Acidovorax caeni sp. nov., a denitrifying species with genetically diverse isolates from activated sludge. Int J Syst Evol Microbiol 58:73–77. https://doi.org/10.1099/ijs.0.65387-0
Holman DB, Brunelle BW, Trachsel J, Allen HK (2017) Meta-analysis to define a core microbiota in the swine gut. mSystems 2:1–14. https://doi.org/10.1128/msystems.00004-17
Ibarbalz FM, Figuerola ELM, Erijman L (2013) Industrial activated sludge exhibit unique bacterial community composition at high taxonomic ranks. Water Res 47:3854–3864. https://doi.org/10.1016/j.watres.2013.04.010
Ji J, Kakade A, Zhang R et al (2019) Alcohol ethoxylate degradation of activated sludge is enhanced by bioaugmentation with Pseudomonas sp. LZ-B. Ecotoxicol Environ Saf. https://doi.org/10.1016/j.ecoenv.2018.11.045
Jiang B, Zhou Z, Dong Y et al (2015a) Bioremediation of petrochemical wastewater containing BTEX compounds by a new immobilized bacterium Comamonas sp. JB in magnetic gellan gum. Appl Biochem Biotechnol 176:572–581. https://doi.org/10.1007/s12010-015-1596-0
Jiang B, Zhou Z, Dong Y et al (2015) Biodegradation of benzene, toluene, ethylbenzene, and o-, m-, and p-xylenes by the newly isolated bacterium Comamonas sp. JB. Appl Biochem Biotechnol. https://doi.org/10.1007/s12010-015-1671-6
Kachouri F, Hamdi M (2004) Enhancement of polyphenols in olive oil by contact with fermented olive mill wastewater by Lactobacillus plantarum. Process Biochem. https://doi.org/10.1016/S0032-9592(03)00189-4
Kim E, Lee J, Han G, Hwang S (2018) Comprehensive analysis of microbial communities in full-scale mesophilic and thermophilic anaerobic digesters treating food waste-recycling wastewater. Bioresour Technol 259:442–450
Kittelmann S, Seedorf H, Walters WA et al (2013) Simultaneous amplicon sequencing to explore Co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in Rumen microbial communities. PLoS ONE. https://doi.org/10.1371/journal.pone.0047879
Kodama Y, Ha LT, Watanabe K (2007) Sulfurospirillum cavolei sp. Nov., a facultatively anaerobic sulfur-reducing bacterium isolated from an underground crude oil storage cavity. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijs.0.64823-0
Lamendella R, Santo Domingo JW, Kelty C, Oerther DB (2008) Bifidobacteria in feces and environmental waters. Appl Environ Microbiol. https://doi.org/10.1128/AEM.01221-07
Lee KC, Darah I, Ibrahim CO (2011) A laboratory scale bioremediation of Tapis crude oil contaminated soil by bioaugmentation of Acinetobacter baumannii T30C. Afr J Microbiol Res. https://doi.org/10.5897/ajmr11.185
Levican A, Collado L, Figueras MJ (2013) Arcobacter cloacae sp. nov., and Arcobacter suis sp. nov., two new species isolated from food and sewage. Syst Appl Microbiol. https://doi.org/10.1016/j.syapm.2012.11.003
Lisle JT, Smith JJ, Edwards DD, McFeters GA (2004) Occurrence of microbial indicators and Clostridium perfringens in wastewater, water column samples, sediments, drinking water, and Weddell seal feces collected at McMurdo Station, Antarctica. Appl Environ Microbiol. https://doi.org/10.1128/AEM.70.12.7269-7276.2004
Liu B, Zhang F, Feng X et al (2006) Thauera and Azoarcus as functionally important genera in a denitrifying quinoline-removal bioreactor as revealed by microbial community structure comparison. FEMS Microbiol Ecol. https://doi.org/10.1111/j.1574-6941.2005.00033.x
Liu J, Yu Y, Chang Y et al (2016a) Enhancing quinoline and phenol removal by adding Comamonas testosteroni bdq06 in treatment of an accidental dye wastewater. Int Biodeterior Biodegrad 115:74–82. https://doi.org/10.1016/j.ibiod.2016.07.016
Liu J, Yu Y, Chang Y et al (2016b) Enhancing quinoline and phenol removal by adding Comamonas testosteroni bdq06 in treatment of an accidental dye wastewater. Int Biodeterior Biodegrad. https://doi.org/10.1016/j.ibiod.2016.07.016
Liu T, Ahn H, Sun W et al (2016c) Identification of a Ruminococcaceae Species as the methyl tert-butyl ether (MTBE) degrading bacterium in a Methanogenic Consortium. Environ Sci Technol. https://doi.org/10.1021/acs.est.5b04731
Liu YX, Hu T, Song Y et al (2015) Heterotrophic nitrogen removal by Acinetobacter sp. Y1 isolated from coke plant wastewater. J Biosci Bioeng. https://doi.org/10.1016/j.jbiosc.2015.03.015
Liu Z, Xie W, Li D et al (2016d) Biodegradation of phenol by bacteria strain Acinetobacter calcoaceticus PA isolated from phenolic wastewater. Int J Environ Res Public Health. https://doi.org/10.3390/ijerph13030300
Lood R, Ertürk G, Mattiasson B (2017) Revisiting antibiotic resistance spreading in wastewater treatment plants– bacteriophages as a much neglected potential transmission vehicle. Front Microbiol 8:1–7. https://doi.org/10.3389/fmicb.2017.02298
Lu H, Chandran K, Stensel D (2014) Microbial ecology of denitrification in biological wastewater treatment. Water Res 64:237–254
Lueders T, Muyzer G (2016) The ecology of anaerobic degraders of BTEX hydrocarbons in aquifers. FEMS Microbiol Ecol 93(1):fiw220
Lünsmann V, Kappelmeyer U, Benndorf R et al (2016) In situprotein-SIP highlights Burkholderiaceae as key players degrading toluene by para ring hydroxylation in a constructed wetland model. Environ Microbiol. https://doi.org/10.1111/1462-2920.13133
Ma J, Wang Z, Zang L et al (2015a) Occurrence and fate of potential pathogenic bacteria as revealed by pyrosequencing in a full-scale membrane bioreactor treating restaurant wastewater. RSC Adv 5:24469–24478. https://doi.org/10.1039/c4ra10220g
Ma Q, Qu Y, Shen W et al (2015b) Bacterial community compositions of coking wastewater treatment plants in steel industry revealed by Illumina high-throughput sequencing. Bioresour Technol 179:436–443. https://doi.org/10.1016/j.biortech.2014.12.041
Magot M, Ravot G, Campaignolle X, Ollivier B, Patel BKC, Fardeau M-L, Thomas P, Crolet J-L, Garcia J-L (1997) Dethiosulfovibrio peptidovorans gen. nov., sp. nov., a New Anaerobic, Slightly Halophilic, Thiosulfate-Reducing Bacterium from Corroding Offshore Oil Wells. Int J Syst Bacteriol 47(3):818–824
Malik A, Jaiswal R (2000) Metal resistance in Pseudomonas strains isolated from soil treated with industrial wastewater. World J Microbiol Biotechnol 16:177–182. https://doi.org/10.1023/A:1008905902282
Martins M, Sanches S, Pereira IAC (2018) Anaerobic biodegradation of pharmaceutical compounds: new insights into the pharmaceutical-degrading bacteria. J Hazard Mater. https://doi.org/10.1016/j.jhazmat.2018.06.001
McMurdie PJ, Holmes S (2013) Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. https://doi.org/10.1371/journal.pone.0061217
Meerbergen K, Van Geel M, Waud M et al (2017) Assessing the composition of microbial communities in textile wastewater treatment plants in comparison with municipal wastewater treatment plants. Microbiologyopen 6:1–13. https://doi.org/10.1002/mbo3.413
Milaković M, Vestergaard G, González-Plaza JJ et al (2019) Pollution from azithromycin-manufacturing promotes macrolide-resistance gene propagation and induces spatial and seasonal bacterial community shifts in receiving river sediments. Environ Int 123:501–511. https://doi.org/10.1016/j.envint.2018.12.050
Muller EEL, Hourcade E, Louhichi-Jelail Y et al (2011) Functional genomics of dichloromethane utilization in Methylobacterium extorquens DM4. Environ Microbiol. https://doi.org/10.1111/j.1462-2920.2011.02524.x
Nagpal S, Haque MM, Singh R, Mande SS (2019) IVikodak-A platform and standard workflow for inferring, analyzing, comparing, and visualizing the functional potential of microbial communities. Front Microbiol. https://doi.org/10.3389/fmicb.2018.03336
Nelson MC, Morrison M, Yu Z (2011) A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour Technol 102:3730–3739. https://doi.org/10.1016/j.biortech.2010.11.119
Ng C, Tan B, Jiang X et al (2019) Metagenomic and resistome analysis of a full-scale municipal wastewater treatment plant in Singapore containing membrane bioreactors. Front Microbiol 10:1–13. https://doi.org/10.3389/fmicb.2019.00172
Niemi RM, Niemela SI, Bamford DH et al (1993) Presumptive fecal streptococci in environmental samples characterized by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Appl Environ Microbiol. https://doi.org/10.1128/aem.59.7.2190-2196.1993
Paradis E (2012) Analysis of phylogenetics and evolution with R, 2nd edn. Springer, Cham
Peng Q, Jiang S, Chen J et al (2018) Unique microbial diversity and metabolic pathway features of fermented vegetables from Hainan, China. Front Microbiol 9:1–12. https://doi.org/10.3389/fmicb.2018.00399
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2012) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(D1):D590–D596
Rajilić-Stojanović M, de Vos WM (2014) The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. https://doi.org/10.1111/1574-6976.12075
Ramadass K, Megharaj M, Venkateswarlu K, Naidu R (2018) Bioavailability of weathered hydrocarbons in engine oil-contaminated soil: impact of bioaugmentation mediated by Pseudomonas spp. on bioremediation. Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2018.04.379
Ritchie H, Roser M (2019) Clean Water. https://ourworldindata.org/water-access. Accessed 21 June 2020
Rizzo L, Manaia C, Merlin C et al (2013) Science of the total environment urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ 447:345–360. https://doi.org/10.1016/j.scitotenv.2013.01.032
Santiago VM, Oliveira VM, Rodrigues MV et al (2012) Phylogenetic and functional diversity of metagenomic libraries of phenol degrading sludge from petroleum refinery wastewater treatment system. AMB Express 2:18. https://doi.org/10.1186/2191-0855-2-18
Sarkar B, Chakrabarti PP, Vijaykumar A, Kale V (2006) Wastewater treatment in dairy industries - possibility of reuse. Desalination 195:141–152. https://doi.org/10.1016/j.desal.2005.11.015
Sarkar P, Roy A, Pal S et al (2017) Enrichment and characterization of hydrocarbon-degrading bacteria from petroleum refinery waste as potent bioaugmentation agent for in situ bioremediation. Bioresour Technol. https://doi.org/10.1016/j.biortech.2017.05.010
Seesuriyachan P, Kuntiya A, Sasaki K, Techapun C (2009) Biocoagulation of dairy wastewater by Lactobacillus casei TISTR 1500 for protein recovery using micro-aerobic sequencing batch reactor (micro-aerobic SBR). Process Biochem. https://doi.org/10.1016/j.procbio.2008.12.006
Selvarajan R, Sibanda T, Venkatachalam S et al (2018) Industrial wastewaters harbor a unique diversity of bacterial communities revealed by high-throughput amplicon analysis. Ann Microbiol 68:445–458. https://doi.org/10.1007/s13213-018-1349-8
Shakibaie MR, Kapadnis BP, Dhakephalker P, Chopade BA (2011) Removal of silver from photographic wastewater effluent using Acinetobacter baumannii BL54. Can J Microbiol 45:995–1000. https://doi.org/10.1139/w99-077
Shchegolkova NM, Krasnov GS, Belova AA et al (2016) Microbial community structure of activated sludge in treatment plants with different wastewater compositions. Front Microbiol 7:1–15. https://doi.org/10.3389/fmicb.2016.00090
Sigala J, Stringam B, Unc A (2017) In vitro examination of the application of saline concentrate to septic tank wastewater. Sustain Water Resour Manag 3:157–162. https://doi.org/10.1007/s40899-017-0089-4
Singh C, Lin J (2010) Bioaugmentation efficiency of diesel degradation by Bacillus pumilus JLB and Acinetobacter calcoaceticus LT1 in contaminated soils. Afr J Biotechnol. https://doi.org/10.4314/ajb.v9i41
Singleton DR, Jones MD, Richardson SD, Aitken MD (2013) Pyrosequence analyses of bacterial communities during simulated in situ bioremediation of polycyclic aromatic hydrocarbon-contaminated soil. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-012-4531-0
Song Z, Edwards SR, Burns RG (2005) Biodegradation of naphthalene-2-sulfonic acid present in tannery wastewater by bacterial isolates Arthrobacter sp. 2AC and Comamonas sp. 4BC. Biodegradation 16:237–252. https://doi.org/10.1007/s10532-004-0889-8
Song Z, Edwards SR, Burns RG (2006) Treatment of naphthalene-2-sulfonic acid from tannery wastewater by a granular activated carbon fixed bed inoculated with bacterial isolates Arthrobacter globiformis and Comamonas testosteroni. Water Res. https://doi.org/10.1016/j.watres.2005.11.035
Stanier RY, Palleroni NJ, Doudoroff M (1966) The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. https://doi.org/10.1099/00221287-43-2-159
Stokholm-Bjerregaard M, McIlroy SJ, Nierychlo M et al (2017) A critical assessment of the microorganisms proposed to be important to enhanced biological phosphorus removal in full-scale wastewater treatment systems. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00718
Su JJ, Yeh KS, Tseng PW (2006) A strain of Pseudomonas sp. isolated from piggery wastewater treatment systems with heterotrophic nitrification capability in Taiwan. Curr Microbiol 53:77–81. https://doi.org/10.1007/s00284-006-0021-x
Szekeres E, Baricz A, Chiriac CM et al (2017) Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ Pollut. https://doi.org/10.1016/j.envpol.2017.01.054
Tang H, Zhang Y, Hu J et al (2019) Mixture of different Pseudomonas aeruginosa SD-1 strains in the efficient bioaugmentation for synthetic livestock wastewater treatment. Chemosphere. https://doi.org/10.1016/j.chemosphere.2019.124455
Tao W, Zhang XX, Zhao F et al (2016) High levels of antibiotic resistance genes and their correlations with bacterial community and mobile genetic elements in pharmaceutical wastewater treatment bioreactors. PLoS ONE 11:1–17. https://doi.org/10.1371/journal.pone.0156854
Tian S, Tian Z, Yang H et al (2017) Detection of viable bacteria during sludge ozonation by the combination of ATP assay with PMA-miseq sequencing. Water (Switzerland). https://doi.org/10.3390/w9030166
Tondee T, Sirianuntapiboon S (2008) Decolorization of molasses wastewater by Lactobacillus plantarum No. PV71-1861. Bioresour Technol 99:6258–6265. https://doi.org/10.1016/j.biortech.2007.12.028
Tyagi M, da Fonseca MMR, de Carvalho CCCR (2011) Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes. Biodegradation 22(2):231–241
Vandewalle JL, Goetz GW, Huse SM et al (2012) Acinetobacter, Aeromonas and Trichococcus populations dominate the microbial community within urban sewer infrastructure. Environ Microbiol. https://doi.org/10.1111/j.1462-2920.2012.02757.x
Vaz-Moreira I, Novo A, Hantsis-Zacharov E et al (2011) Acinetobacter rudis sp. nov., isolated from raw milk and raw wastewater. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijs.0.027045-0
Walden C, Carbonero F, Zhang W (2017) Assessing impacts of DNA extraction methods on next generation sequencing of water and wastewater samples. J Microbiol Methods 141:10–16
Wang CC, Lee CM, Chen LJ (2004) Removal of nitriles from synthetic wastewater by acrylonitrile utilizing bacteria. J Environ Sci Health Part A Toxic/Hazard Subst Environ Eng 39(7):1767–1779
Wang M, Xu J, Wang J et al (2013) Differences between 4-fluoroaniline degradation and autoinducer release by Acinetobacter sp. TW: implications for operating conditions in bacterial bioaugmentation. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-013-1660-7
Wang R, Han S, Sun C et al (2016) Seohaeicola zhoushanensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.001140
Weelink SAB, van Eekert MHA, Stams AJM (2010) Degradation of BTEX by anaerobic bacteria: physiology and application. Rev Environ Sci Bio/Technol 9(4):359–385
Wéry N, Monteil C, Pourcher AM, Godon JJ (2010) Human-specific fecal bacteria in wastewater treatment plant effluents. Water Res. https://doi.org/10.1016/j.watres.2009.11.027
Wickham H (2016) ggplot2 elegant graphics for data analysis. Springer, Cham
Xu LH, Tan Z, Zhang C et al (2017) Performance and microbial diversity of a full-scale oilfield wastewater treatment plant. Desalin Water Treat 99:239–247. https://doi.org/10.5004/dwt.2017.21694
Yan P, Wang J, Chen YP et al (2015) Investigation of microbial community structure in an advanced activated sludge side-stream reactor process with alkaline treatment. Int Biodeterior Biodegrad. https://doi.org/10.1016/j.ibiod.2015.07.003
Yang L, Cai T, Ding D et al (2017) Biodegradation of 2-hydroxyl-1,4 naphthoquinone (lawsone) by Pseudomonas taiwanensis LH-3 isolated from activated sludge. Sci Rep. https://doi.org/10.1038/s41598-017-06338-1
Yang L, Ren YX, Liang X et al (2015) Nitrogen removal characteristics of a heterotrophic nitrifier Acinetobacter junii YB and its potential application for the treatment of high-strength nitrogenous wastewater. Bioresour Technol. https://doi.org/10.1016/j.biortech.2015.05.075
Zakaria ZA, Zakaria Z, Surif S, Ahmad WA (2007) Hexavalent chromium reduction by Acinetobacter haemolyticus isolated from heavy-metal contaminated wastewater. J Hazard Mater. https://doi.org/10.1016/j.jhazmat.2006.11.052
Zhang J, Kobert K, Flouri T, Stamatakis A (2014) PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. https://doi.org/10.1093/bioinformatics/btt593
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sector.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Editorial responsibility: Gobinath Ravindran.
Rights and permissions
About this article
Cite this article
Palanisamy, V., Gajendiran, V. & Mani, K. Meta-analysis to identify the core microbiome in diverse wastewater. Int. J. Environ. Sci. Technol. 19, 5079–5096 (2022). https://doi.org/10.1007/s13762-021-03349-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-021-03349-4