Abstract
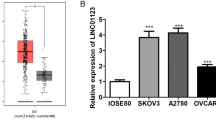
Epithelial Ovarian Cancer (EOC) is a heterogeneous disease usually diagnosed at advanced stages. Therefore, early detection is crucial for better survival. Despite the advances in ovarian research, mechanisms underlying EOC carcinogenesis are not elucidated. We performed chromatin immunoprecipitation sequencing to identify genes regulated by E2F5, a transcription factor involved in ovarian carcinogenesis. Results revealed several putative candidate genes (115 protein-coding genes, 20 lncRNAs, 6 pseudogenes, and 4 miRNAs). A literature review and bioinformatics analysis of these genes revealed a novel lncRNA candidate (LINC01465) in EOC. We validated LINC01465 by quantifying its expression in EOC cell lines and selected OVSAHO and SKOV3 as a model with high LINC01465 levels. We silenced LINC01465 and performed proliferation, wound healing, invasion, and drug resistance assays. Knocking-down LINC01465 resulted in reduced migration, suggesting potential involvement in EOC. Furthermore, to identify the significance of LINC01465 in chemoresistance, we assessed the LINC01465 levels in A2780 S cells treated with malformin, which revealed higher LINC01465 expression as compared to untreated A2780S cells implying the involvement of LINC01465 in cell death. Thus, this study unraveled the repertoire of E2F5 regulated candidate genes and suggested a putative role of LINC01465 in malformin-induced cell death in EOC.










Similar content being viewed by others
Data availability statement
The data sets generated during and/or analyzed during this study are included in this published article.
References
Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018: Ovarian Cancer Statistics, 2018. CA Cancer J Clin. 2018;68:284–96.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Howlader N, Noone A, Krapcho M, Garshell J, Miller D, Altekruse S, et al. SEER cancer statistics review, 1975–2012. 2014;
Zhang B, Cai FF, Zhong XY. An overview of biomarkers for the ovarian cancer diagnosis. Eur J Obstet Gynecol Reprod Biol. 2011;158:119–23.
Kothandaraman N, Bajic VB, Brendan PN, Huak CY, Keow PB, Razvi K, et al. E2F5 status significantly improves malignancy diagnosis of epithelial ovarian cancer. BMC Cancer. 2010;10:64. https://doi.org/10.1186/1471-2407-10-64.
Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–56.
Liban TJ, Thwaites MJ, Dick FA, Rubin SM. Structural conservation and E2F binding specificity within the retinoblastoma pocket protein family. J Mol Biol. 2016;428:3960–71.
Gaubatz S, Lindeman GJ, Ishida S, Jakoi L, Nevins JR, Livingston DM, et al. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell. 2000;6:729–35.
Lu G, Sun Y, An S, Xin S, Ren X, Zhang D, et al. MicroRNA-34a targets FMNL2 and E2F5 and suppresses the progression of colorectal cancer. Exp Mol Pathol. 2015;99:173–9.
Zhao J, Wu X-Y, Ling X-H, Lin Z-Y, Fu X, Deng Y-H, et al. Analysis of genetic aberrations on chromosomal region 8q21-24 identifies E2F5 as an oncogene with copy number gain in prostate cancer. Med Oncol Northwood Lond Engl. 2013;30:465.
Zou C, Li Y, Cao Y, Zhang J, Jiang J, Sheng Y, et al. Up-regulated MicroRNA-181a induces carcinogenesis in hepatitis B virus-related hepatocellular carcinoma by targeting E2F5. BMC Cancer. 2014;14:97.
Ishimoto T, Shiozaki A, Ichikawa D, Fujiwara H, Konishi H, Komatsu S, et al. E2F5 as an independent prognostic factor in esophageal squamous cell carcinoma. Anticancer Res. 2013;33:5415–20.
Tian H, Hou L, Xiong Y-M, Huang J-X, Zhang W-H, Pan Y-Y, et al. miR-132 targeting E2F5 suppresses cell proliferation, invasion, migration in ovarian cancer cells. Am J Transl Res. 2016;8(3):1492–501.
Malgundkar SH, Burney I, Al Moundhri M, Al Kalbani M, Lakhtakia R, Okamoto A, et al. FAT4 silencing promotes epithelial-to-mesenchymal transition and invasion via regulation of YAP and β-catenin activity in ovarian cancer. BMC Cancer. 2020. https://doi.org/10.1186/s12885-020-06900-7.
Shridhar V, Lee J, Pandita A. Genetic analysis of early- versus late-stage ovarian tumors. Cancer Res. 2001;61(15):5895–904.
Wei J-W, Huang K, Yang C, Kang C-S. Non-coding RNAs as regulators in epigenetics. Oncol Rep. 2017;37:3–9.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41.
Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Zamaraev AV, Volik PI, Sukhikh GT, Kopeina GS, Zhivotovsky B. Long non-coding RNAs: a view to kill ovarian cancer. Biochim Biophys Acta BBA. 2021;1876: 188584.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Wery M, Kwapisz M, Morillon A. Noncoding RNAs in gene regulation. WIREs Syst Biol Med. 2011;3:728–38.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146:353–8.
Zhou R-S, Zhang E-X, Sun Q-F, Ye Z-J, Liu J-W, Zhou D-H, et al. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer. 2019;19:779.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407.
Yu C, Sun J, Leng X, Yang J. Long noncoding RNA SNHG6 functions as a competing endogenous RNA by sponging miR-181a-5p to regulate E2F5 expression in colorectal cancer. Cancer Manag Res. 2019;11:611–24.
Wang Y, Wang X, Han L, Hu D. LncRNA MALAT1 regulates the progression and cisplatin resistance of ovarian cancer cells via Modulating miR-1271-5p/E2F5 Axis. Cancer Manag Res. 2020;12:9999–10010.
Wang L, Yu M, Zhao S. lncRNA MEG3 modified epithelial-mesenchymal transition of ovarian cancer cells by sponging miR-219a-5p and regulating EGFR. J Cell Biochem. 2019;120:17709–22.
Patterson AD, Gonzalez FJ, Perdew GH, Peters JM. Molecular regulation of carcinogenesis: friend and foe. Toxicol Sci. 2018;165:277–83.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–26.
Gorai I, Nakazawa T, Miyagi E, Hirahara F, Nagashima Y, Minaguchi H. Establishment and characterization of two human ovarian clear cell adenocarcinoma lines from metastatic lesions with different properties. Gynecol Oncol. 1995;57(1):33–46. https://doi.org/10.1006/gyno.1995.1097. (PMID: 7535723).
Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013. https://doi.org/10.1038/ncomms3126.
Anglesio MS, Wiegand KC, Melnyk N, Chow C, Salamanca C, Prentice LM, et al. Type-specific cell line models for type-specific ovarian cancer research. PLoS ONE. 2013;8:e72162.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–8.
Abdullah N, Tamimi Y, Dobretsov S, Balushi NA, Alshekaili J, Al Balushi H, et al. Malformin-A1 (MA1) sensitizes chemoresistant ovarian cancer cells to cisplatin-induced apoptosis. Mol Basel Switz. 2021;26:3624.
Zheng Z, Li X, You H, Zheng X, Ruan X. LncRNA SOCS2-AS1 inhibits progression and metastasis of colorectal cancer through stabilizing SOCS2 and sponging miR-1264. Aging. 2020;12:10517–26.
Li S-M, Wu H-L, Yu X, Tang K, Wang S-G, Ye Z-Q, et al. The putative tumour suppressor miR-1-3p modulates prostate cancer cell aggressiveness by repressing E2F5 and PFTK1. J Exp Clin Cancer Res. 2018. https://doi.org/10.1186/s13046-018-0895-z.
Huang J, Lin F, Xu C, Xu Y. LINC00662 facilitates osteosarcoma progression via sponging miR-103a-3p and regulating SIK2 expression. J Tissue Eng Regen Med. 2021;15:1082–91.
Zhai Y, Liu Y, Wang Z, Wang W, Zhou J, Lu J. Long non-coding RNA LINC00313 accelerates cervical carcinoma progression by miR-4677-3p/CDK6 axis. OncoTargets Ther. 2021;14:2213–26.
da Conceição IMCA, Luscher-Dias T, Queiroz LR, de Melo AGB, Machado CR, Gomes KB, et al. Metformin treatment modulates long non-coding RNA isoforms expression in human cells. Non-Coding RNA. 2022;8:68.
Wang B-G, Jiang L-Y, Xu Q. A comprehensive evaluation for polymorphisms in let-7 family in cancer risk and prognosis: a system review and meta-analysis. 2018. Biosci Rep. https://doi.org/10.1042/BSR20180273.
Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68:10307–14.
Funding
This research was funded by the grant from His Majesty Trust Fund (SR/MED/BIOC/14/01).
Author information
Authors and Affiliations
Contributions
YT participated in the conceptualization and designed the project methodology. SH, NA, HB, IG, ZH, and HB performed the experiments. SH interpreted the data and wrote the manuscript under the supervision of YT. AO, IB, MK, HB, and RL contributed to discussing and reviewing the clinical part of the project.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Malgundkar, S.H., Hassan, N.A., Al Badi, H. et al. Identification and validation of a novel long non-coding RNA (LINC01465) in ovarian cancer. Human Cell 36, 762–774 (2023). https://doi.org/10.1007/s13577-022-00842-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-022-00842-x