Abstract
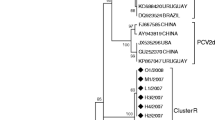
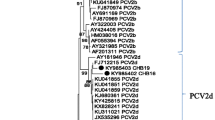
This study aimed to investigate the co-infection and genetic characteristics of Porcine circoviruses in PMWS-affected pigs in five commercial farrow-to-finish swine farms in Vietnam. By the end of 2022, the percentage of PMWS-affected pigs in these farms has increased significantly compared to previous years. The lymph node samples from ten PMWS typical cases were randomly collected to test for the presence of PRRSV, PCV2, PCV3 and PCV4. While PRRSV and PCV4 were not found in these cases, 10 and 3 out of 10 samples were positive for PCV2 and PCV3, respectively. Three farms in the study showed the co-infection of PCV2 and PCV3 in affected pigs. Besides, all PCV-positive samples were sequenced to evaluate genetic characterization of PCVs in PMWS-affected cases. Phylogenetic analysis showed that all PCV3 strains in the study were clustered into PCV3b genotype. 8 out of 10 PCV2 strains belonged to PCV2d genotype while the remaining two strains belonged to PCV2b genotypes. Two farms had co-circulation of PCV2b and PCV2d genotypes in two different age groups of pigs, which is reported for the first time in Vietnam. Several amino acid substitutions were identified in important antigenic regions in the capsid protein of the PCV2 field strains compared to vaccine strains. Taken together, the results showed the high co-prevalence of PCV3 and PCV2, and the wide genetic diversity of PCV2 field and vaccine strains may be the cause of the increased PMWS situation in these pig farms.


Similar content being viewed by others
Data availability
The complete genome sequences of ten PCV2 strains and the ORF2 genes of three PCV3 strains in this study have been deposited in GenBank under the accession numbers OQ784905-14 and OR058801-03, respectively.
References
Afghah Z, Webb B, Meng X-J, Ramamoorthy S. Ten years of PCV2 vaccines and vaccination: Is eradication a possibility? Vet Microbiol. 2017;206:21–8.
Arruda B, Piñeyro P, Derscheid R, Hause B, Byers E, Dion K, Long D, Sievers C, Tangen J, Williams T. PCV3-associated disease in the United States swine herd. Emerg Microbes Infect. 2019;8:684–98.
Chae C. A review of porcine circovirus 2-associated syndromes and diseases. Vet J. 2005;169:326–36.
Chen G, Mai K, Zhou L, Wu R, Tang X, Wu J, He L, Lan T, Xie Q, Sun Y. Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transbound Emerg Dis. 2017;64:1650–4.
Chen N, Huang Y, Ye M, Li S, Xiao Y, Cui B, Zhu J. Co-infection status of classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circoviruses (PCV2 and PCV3) in eight regions of China from 2016 to 2018. Infect Genet Evol. 2019;68:127–35.
Constans M, Ssemadaali M, Kolyvushko O, Ramamoorthy S. Antigenic determinants of possible vaccine escape by porcine circovirus subtype 2b viruses. Bioinform Biol Insights. 2015;9:BBI-S30226.
D’silva AL, Bharali A, Buragohain L, Pathak DC, Ramamurthy N, Batheja R, Mariappan AK, Gogoi SM, Barman NN, Dey S. Molecular characterization of porcine circovirus 2 circulating in Assam and Arunachal Pradesh of India. Anim Biotechnol. 2023;34:462–6.
de Boisseson C, Beven V, Bigarre L, Thiery R, Rose N, Eveno E, Madec F, Jestin A. Molecular characterization of porcine circovirus type 2 isolates from post-weaning multisystemic wasting syndrome-affected and non-affected pigs. J Gen Virol. 2004;85:293–304.
Dinh PX, Nguyen HN, Lai DC, Nguyen TT, Nguyen NM, Do DT. Genetic diversity in the capsid protein gene of porcine circovirus type 3 in Vietnam from 2018 to 2019. Arch Virol. 2023;168:30.
Dinh PX, Nguyen MN, Nguyen HT, Tran VH, Tran QD, Dang KH, Vo DT, Le HT, Nguyen NTT, Nguyen TT. Porcine circovirus genotypes and their copathogens in pigs with respiratory disease in southern provinces of Vietnam. Arch Virol. 2021;166:403–11.
Dinh XP, Nguyen TTT, Vu LXH, Tran KT. Identification of porcine circovirus type 2 (PCV2), type 3 (PCV3) and porcine parvovirus (PPV) in swine by multiplex PCR test. J Agric Dev. 2021;20:11–7.
Doan HTT, Do RT, Thao PTP, Le XTK, Nguyen KT, Hien NTT, Duc LM, Pham LTK, Le TH. Molecular genotypic analysis of porcine circovirus type 2 reveals the predominance of PCV2d in Vietnam (2018–2020) and the association between PCV2h, the recombinant forms, and Vietnamese vaccines. Arch Virol. 2022;167:2011–26.
Fenaux M, Halbur PG, Gill M, Toth TE, Meng X-J. Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2. J Clin Microbiol. 2000;38:2494–503.
Ge M, Yan A, Luo W, Hu Y-F, Li R-C, Jiang D-L, Yu X-L. Epitope screening of the PCV2 Cap protein by use of a random peptide-displayed library and polyclonal antibody. Virus Res. 2013;177:103–7.
Guo J, Hou L, Zhou J, Wang D, Cui Y, Feng X, Liu J. Porcine circovirus type 2 vaccines: Commercial application and research advances. Viruses. 2022;14:2005.
Guo L, Lu Y, Huang L, Wei Y, Liu C. Identification of a new antigen epitope in the nuclear localization signal region of porcine circovirus type 2 capsid protein. Intervirology. 2011;54:156–63.
Guo Z, Ruan H, Qiao S, Deng R, Zhang G. Co-infection status of porcine circoviruses (PCV2 and PCV3) and porcine epidemic diarrhea virus (PEDV) in pigs with watery diarrhea in Henan province, central China. Microb Pathog. 2020;142:104047.
Huynh T, Nguyen B, Nguyen V, Dang H, Mai T, Tran T, Ngo M, Le V, Vu T, Ta T. Phylogenetic and Phylogeographic analyses of porcine circovirus type 2 among pig farms in Vietnam. Transbound Emerg Dis. 2014;61:e25–34.
Jiang H, Wang D, Wang J, Zhu S, She R, Ren X, Tian J, Quan R, Hou L, Li Z. Induction of porcine dermatitis and nephropathy syndrome in piglets by infection with porcine circovirus type 3. J Virol. 2019;93:e02045-e2118.
Karuppannan AK, Opriessnig T. Porcine circovirus type 2 (PCV2) vaccines in the context of current molecular epidemiology. Viruses. 2017;9:99.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547.
Li W, Li J, Dai X, Liu M, Khalique A, Wang Z, Zeng Y, Zhang D, Ni X, Zeng D, Bo J, Kangcheng P. Surface display of porcine circovirus type 2 antigen protein cap on the spores of bacillus subtilis 168: An effective mucosal vaccine candidate. Front Immunol. 2022;13:5459.
Li X, Chen S, Niu G, Zhang X, Ji W, Ren Y, Zhang L, Ren L. Porcine circovirus type 4 strains circulating in China are relatively stable and have higher homology with mink circovirus than other porcine circovirus types. Int J Mol Sci. 2022;23:3288.
Mahe D, Blanchard P, Truong C, Arnauld C, Le Cann P, Cariolet R, Madec F, Albina E, Jestin A. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J Gen Virol. 2000;81:1815–24.
Nguyen NH, Do Tien D, Nguyen TQ, Nguyen TT, Nguyen MN. Identification and whole-genome characterization of a novel Porcine Circovirus 3 subtype b strain from swine populations in Vietnam. Virus Genes. 2021;57:385–9.
Nguyen V, Chung H, Huynh T, Cao T, Vu T, Le V, Pham H. Molecular characterization of novel Porcine circovirus 3 (PCV3) in pig populations in the North of Vietnam. Arch Gene Genome Res. 2018;1:9.
Nguyen VG, Do HQ, Huynh TML, Park YH, Park BK, Chung HC. Molecular-based detection, genetic characterization and phylogenetic analysis of porcine circovirus 4 from Korean domestic swine farms. Transbound Emerg Dis. 2022;69:538–48.
Opriessnig T, O’Neill K, Gerber PF, de Castro AM, Gimenéz-Lirola LG, Beach NM, Zhou L, Meng X-J, Wang C, Halbur PG. A PCV2 vaccine based on genotype 2b is more effective than a 2a-based vaccine to protect against PCV2b or combined PCV2a/2b viremia in pigs with concurrent PCV2, PRRSV and PPV infection. Vaccine. 2013;31:487–94.
Qi S, Su M, Guo D, Li C, Wei S, Feng L, Sun D. Molecular detection and phylogenetic analysis of porcine circovirus type 3 in 21 Provinces of China during 2015–2017. Transbound Emerg Dis. 2019;66:1004–15.
Rosario K, Breitbart M, Harrach B, Segalés J, Delwart E, Biagini P, Varsani A. Revisiting the taxonomy of the family Circoviridae: Establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol. 2017;162:1447–63.
Salgado RL, Vidigal PM, de Souza LF, Onofre TS, Gonzaga NF, Eller MR, Bressan GC, Fietto JL, Almeida MR, Silva JA. Identification of an emergent porcine circovirus-2 in vaccinated pigs from a Brazilian farm during a postweaning multisystemic wasting syndrome outbreak. Genome Announc. 2014;2:e00163-e214.
Segalés J. Porcine circovirus type 2 (PCV2) infections: Clinical signs, pathology and laboratory diagnosis. Virus Res. 2012;164:10–9.
Seo HW, Park C, Kang I, Choi K, Jeong J, Park S-J, Chae C. Genetic and antigenic characterization of a newly emerging porcine circovirus type 2b mutant first isolated in cases of vaccine failure in Korea. Arch Virol. 2014;159:3107–11.
Shang S-B, Jin Y-L, Jiang X-T, Zhou J-Y, Zhang X, Xing G, He JL, Yan Y. Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol Immunol. 2009;46:327–34.
Sirisereewan C, Thanawongnuwech R, Kedkovid R. Current understanding of the pathogenesis of porcine circovirus 3. Pathogens. 2022;11:64.
Tan CY, Thanawongnuwech R, Arshad SS, Hassan L, Fong MWC, Ooi PT. Genotype shift of Malaysian porcine circovirus 2 (PCV2) from PCV2b to PCV2d within a decade. Animals. 2022;12:1849.
Trible BR, Ramirez A, Suddith A, Fuller A, Kerrigan M, Hesse R, Nietfeld J, Guo B, Thacker E, Rowland RR. Antibody responses following vaccination versus infection in a porcine circovirus-type 2 (PCV2) disease model show distinct differences in virus neutralization and epitope recognition. Vaccine. 2012;30:4079–85.
Vargas-Bermudez DS, Mogollón JD, Jaime J. The Prevalence and Genetic Diversity of PCV3 and PCV2 in Colombia and PCV4 Survey during 2015–2016 and 2018–2019. Pathogens. 2022;11:633.
Wang D, Mai J, Yang Y, Xiao C-T, Wang N. Current knowledge on epidemiology and evolution of novel porcine circovirus 4. Vet Res. 2022;53:1–9.
Wang Y, Noll L, Lu N, Porter E, Stoy C, Zheng W, Liu X, Peddireddi L, Niederwerder M, Bai J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound Emerg Dis. 2020;67:1284–94.
Woźniak A, Miłek D, Bąska P, Stadejek T. Does porcine circovirus type 3 (PCV3) interfere with porcine circovirus type 2 (PCV2) vaccine efficacy? Transbound Emerg Dis. 2019;66:1454–61.
Xiao C, Halbur P, Opriessnig T. Complete genome sequence of a novel porcine circovirus type 2b variant present in cases of vaccine failures in the United States. J Virol. 2012;86:12469–12469.
Zhai SL, Chen SN, Wei ZZ, Zhang JW, Huang L, Lin T, Yue C, Ran DL, Yuan SS, Wei WK. Co-existence of multiple strains of porcine circovirus type 2 in the same pig from China. Virol J. 2011;8:1–5.
Zhan Y, Yu W, Cai X, Lei X, Lei H, Wang A, Sun Y, Wang N, Deng Z, Yang Y. The carboxyl terminus of the porcine circovirus type 2 capsid protein is critical to virus-like particle assembly, cell entry, and propagation. J Virol. 2020;94:e00042-e120.
Zhang L, Luo Y, Liang L, Li J, Cui S. Phylogenetic analysis of porcine circovirus type 3 and porcine circovirus type 2 in China detected by duplex nanoparticle-assisted PCR. Infect Genet Evol. 2018;60:1–6.
Zhang S, Wang D, Jiang Y, Li Z, Zou Y, Li M, Yu H, Huang K, Yang Y, Wang N. Development and application of a baculovirus-expressed capsid protein-based indirect ELISA for detection of porcine circovirus 3 IgG antibodies. BMC Vet Res. 2019;15:79.
Zou Y, Zhang N, Zhang J, Zhang S, Jiang Y, Wang D, Tan Q, Yang Y, Wang N. Molecular detection and sequence analysis of porcine circovirus type 3 in sow sera from farms with prolonged histories of reproductive problems in Hunan. China Arch Virol. 2018;163:2841–7.
Acknowledgements
This work was the study of a project funded by the Ministry of Education and Training, Vietnam (B2023-NLS-04).
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
The study was conducted in compliance with the institutional rules for the care and use of laboratory animals and using a protocol approved by the Ministry of Agriculture and Rural Development (MARD) Vietnam (TCVN 8402:2010).
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lai, D.C., Nguyen, D.M.T., Nguyen, T.T. et al. Co-circulation and genetic characteristics of porcine circoviruses in postweaning multisystemic wasting syndrome cases in commercial swine farms. VirusDis. 34, 531–538 (2023). https://doi.org/10.1007/s13337-023-00849-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-023-00849-4