Abstract
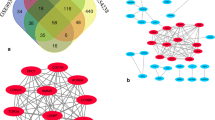
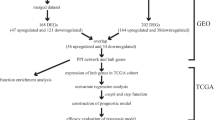
Obesity, which has become one of the biggest public health problems of the twenty-first century, accompanies many chronic conditions, including cancer. On the other hand, liver cancer, which is known to be associated with obesity, is considered another serious threat to public health. However, the underlying drivers of the development of obesity-associated hepatocellular carcinoma (HCC) remain blurry. The current study attempted to identify the key genes and pathways in the obesity-induced development of HCC using integrated bioinformatics analyses. Obesity and HCC-associated gene expression datasets were downloaded from Gene Expression Omnibus (GEO) and analyzed to identify overlapping differentially expressed genes (DEGs) and hub genes. The prognostic potentials, survival analysis, and expression levels of hub genes were further assessed. Moreover, the correlation between hub genes and the immune cells infiltration was analyzed. The findings of this research revealed that both mRNA and protein expression levels of the four hub genes (IGF1, ACADL, CYP2C9, and G6PD) involved in many important metabolic pathways are remarkably altered in both obese individuals and patients with HCC. The results demonstrated that these dysregulated genes in both obesity and HCC may serve as considerable targets for the prevention and treatment of HCC development in obese individuals.





Similar content being viewed by others
Data availability
All relevant datasets in the current study are available in the GEO repository.
References
World Health Organization. Noncommunicable diseases country profiles 2018. Geneva: World Health Organization. 2018. https://www.who.int/nmh/publications/ncd-profiles-2018/en/. Last accessed 29 March 2021.
World Health Organization. Global status report on noncommunicable diseases 2014. World Health Organization. 2014. https://www.who.int/nmh/publications/ncd-status-report-2014/en/. Last accessed 29 March 2021.
Collaboration N C D R F. Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet. 2017;390:2627–42. https://doi.org/10.1016/S0140-6736(17)32129-3.
Saitta C, Pollicino T, Raimondo G. Obesity and liver cancer. Ann Hepatol. 2019;18:810–5. https://doi.org/10.1016/j.aohep.2019.07.004.
Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16:1. https://doi.org/10.4103/jcar.JCar_9_16.
El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. https://doi.org/10.1053/j.gastro.2007.04.061.
Balogh J, Victor D 3rd, Asham EH, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. https://doi.org/10.2147/JHC.S61146.
Tripodi A, Mannucci PM. The coagulopathy of chronic liver disease. N Engl J Med. 2011;365:147–56. https://doi.org/10.1056/NEJMra1011170.
Blonski W, Kotlyar DS, Forde KA. Non-viral causes of hepatocellular carcinoma. World J Gastroenterol. 2010;16:3603–15. https://doi.org/10.3748/wjg.v16.i29.3603.
Sun B, Karin M. Obesity, inflammation, and liver cancer. J Hepatol. 2012;56:704–13. https://doi.org/10.1016/j.jhep.2011.09.020.
Larsson SC, Wolk A. Overweight, obesity and risk of liver cancer: a meta-analysis of cohort studies. Br J Cancer. 2007;97:1005–8. https://doi.org/10.1038/sj.bjc.6603932.
Katanoda K, Matsuda T. Five-year relative survival rate of liver cancer in the USA, Europe and Japan. Jpn J Clin Oncol. 2014;44:302–3. https://doi.org/10.1093/jjco/hyu025.
Zhang C, Peng L, Zhang Y, et al. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med Oncol. 2017;34:101. https://doi.org/10.1007/s12032-017-0963-9.
Lvovs D, Favorova OO, Favorov AV. A polygenic approach to the study of polygenic diseases. Acta Naturae. 2012;4:59–71. https://doi.org/10.32607/20758251-2012-4-3-59-71.
Li J, Zheng L, Uchiyama A, et al. A data mining paradigm for identifying key factors in biological processes using gene expression data. Sci Rep. 2018;8:9083. https://doi.org/10.1038/s41598-018-27258-8.
Pihlajamaki J, Boes T, Kim EY, et al. Thyroid hormone-related regulation of gene expression in human fatty liver. J Clin Endocrinol Metab. 2009;94:3521–9. https://doi.org/10.1210/jc.2009-0212.
Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511. https://doi.org/10.1002/hep.27893.
Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–5. https://doi.org/10.1093/nar/gks1193.
Pathan M, Keerthikumar S, Ang CS, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597–601. https://doi.org/10.1002/pmic.201400515.
Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52. https://doi.org/10.1093/nar/gku1003.
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. https://doi.org/10.1101/gr.1239303.
da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. https://doi.org/10.1038/nprot.2008.211.
da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. https://doi.org/10.1093/nar/gkn923.
Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–60. https://doi.org/10.1093/nar/gkz430.
Gyorffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. https://doi.org/10.1371/journal.pone.0082241.
Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58. https://doi.org/10.1016/j.neo.2017.05.002.
Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–10. https://doi.org/10.1158/0008-5472.CAN-17-0307.
Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. https://doi.org/10.1126/science.1260419.
Qi Z, Yan F, Chen D, et al. Identification of prognostic biomarkers and correlations with immune infiltrates among cGAS-STING in hepatocellular carcinoma. 2020. Biosci Rep. https://doi.org/10.1042/BSR20202603.
Hruby A, Hu FB. The epidemiology of obesity: a big picture. Pharmacoeconomics. 2015;33:673–89. https://doi.org/10.1007/s40273-014-0243-x.
Martin KA, Mani MV, Mani A. New targets to treat obesity and the metabolic syndrome. Eur J Pharmacol. 2015;763:64–74. https://doi.org/10.1016/j.ejphar.2015.03.093.
Mitchell NS, Catenacci VA, Wyatt HR, et al. Obesity: overview of an epidemic. Psychiatr Clin N Am. 2011;34:717–32. https://doi.org/10.1016/j.psc.2011.08.005.
Kelly T, Yang W, Chen CS, et al. Global burden of obesity in 2005 and projections to 2030. Int J Obes (Lond). 2008;32:1431–7. https://doi.org/10.1038/ijo.2008.102.
World Health Organization. International Agency for Research on Cancer. Cancer fact sheets. Liver cancer. https://gco.iarc.fr/today/data/factsheets/cancers/11-Liver-fact-sheet.pdf. Last accessed 29 March 2021.
Polyzos SA, Kountouras J, Mantzoros CS. Obesity and nonalcoholic fatty liver disease: from pathophysiology to therapeutics. Metabolism. 2019;92:82–97. https://doi.org/10.1016/j.metabol.2018.11.014.
Estes C, Razavi H, Loomba R, et al. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018;67:123–33. https://doi.org/10.1002/hep.29466.
Wong RJ, Cheung R, Ahmed A. Nonalcoholic steatohepatitis is the most rapidly growing indication for liver transplantation in patients with hepatocellular carcinoma in the U.S. Hepatology. 2014;59:2188–95. https://doi.org/10.1002/hep.26986.
Wrigley S, Arafa D, Tropea D. Insulin-like growth factor 1: at the crossroads of brain development and aging. Front Cell Neurosci. 2017;11:14. https://doi.org/10.3389/fncel.2017.00014.
Xu GS, Li ZW, Huang ZP, et al. MiR-497-5p inhibits cell proliferation and metastasis in hepatocellular carcinoma by targeting insulin-like growth factor 1. Mol Genet Genomic Med. 2019;7:e00860. https://doi.org/10.1002/mgg3.860.
D’Alessandro R, Refolo MG, Lippolis C, et al. Strong enhancement by IGF1-R antagonists of hepatocellular carcinoma cell migration inhibition by Sorafenib and/or vitamin K1. Cell Oncol (Dordr). 2018;41:283–96. https://doi.org/10.1007/s13402-018-0370-z.
Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–62. https://doi.org/10.1038/nrc2664.
Enguita-German M, Fortes P. Targeting the insulin-like growth factor pathway in hepatocellular carcinoma. World J Hepatol. 2014;6:716–37. https://doi.org/10.4254/wjh.v6.i10.716.
Su WW, Lee KT, Yeh YT, et al. Association of circulating insulin-like growth factor 1 with hepatocellular carcinoma: one cross-sectional correlation study. J Clin Lab Anal. 2010;24:195–200. https://doi.org/10.1002/jcla.20320.
Liu B, Balkwill A, Reeves G, et al. Body mass index and risk of liver cirrhosis in middle aged UK women: prospective study. BMJ. 2010;340:c912. https://doi.org/10.1136/bmj.c912.
Aleksandrova K, Stelmach-Mardas M, Schlesinger S. Obesity and liver cancer. Recent Results Cancer Res. 2016;208:177–98. https://doi.org/10.1007/978-3-319-42542-9_10.
Clemmons DR. Role of insulin-like growth factor iin maintaining normal glucose homeostasis. Horm Res. 2004;62(Suppl 1):77–82. https://doi.org/10.1159/000080763.
Lagana AS, Vitale SG, Nigro A, et al. Pleiotropic actions of peroxisome proliferator-activated receptors (PPARs) in dysregulated metabolic homeostasis, ınflammation and cancer: current evidence and future perspectives. Int J Mol Sci. 2016. https://doi.org/10.3390/ijms17070999.
Lamichane S, Dahal Lamichane B, Kwon SM. Pivotal roles of peroxisome proliferator-activated receptors (PPARs) and their signal cascade for cellular and whole-body energy homeostasis. Int J Mol Sci. 2018. https://doi.org/10.3390/ijms19040949.
Antonosante A, d’Angelo M, Castelli V, et al. The ınvolvement of PPARs in the peculiar energetic metabolism of tumor cells. Int J Mol Sci. 2018. https://doi.org/10.3390/ijms19071907.
Chang WH, Lai AG. The pan-cancer mutational landscape of the PPAR pathway reveals universal patterns of dysregulated metabolism and interactions with tumor immunity and hypoxia. Ann N Y Acad Sci. 2019;1448:65–82. https://doi.org/10.1111/nyas.14170.
Houten SM, Wanders RJ. A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation. J Inherit Metab Dis. 2010;33:469–77. https://doi.org/10.1007/s10545-010-9061-2.
Xie BX, Zhang H, Wang J, et al. Analysis of differentially expressed genes in LNCaP prostate cancer progression model. J Androl. 2011;32:170–82. https://doi.org/10.2164/jandrol.109.008748.
Hill VK, Ricketts C, Bieche I, et al. Genome-wide DNA methylation profiling of CpG islands in breast cancer identifies novel genes associated with tumorigenicity. Cancer Res. 2011;71:2988–99. https://doi.org/10.1158/0008-5472.CAN-10-4026.
Huang D, Li T, Li X, et al. HIF-1-mediated suppression of acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 2014;8:1930–42. https://doi.org/10.1016/j.celrep.2014.08.028.
Zhao X, Qin W, Jiang Y, et al. ACADL plays a tumor-suppressor role by targeting Hippo/YAP signaling in hepatocellular carcinoma. NPJ Precis Oncol. 2020;4:7. https://doi.org/10.1038/s41698-020-0111-4.
Yu D, Green B, Marrone A, et al. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015;5:8534. https://doi.org/10.1038/srep08534.
Chan AT, Tranah GJ, Giovannucci EL, et al. A prospective study of genetic polymorphisms in the cytochrome P-450 2C9 enzyme and the risk for distal colorectal adenoma. Clin Gastroenterol Hepatol. 2004;2:704–12. https://doi.org/10.1016/s1542-3565(04)00294-0.
Martinez C, Garcia-Martin E, Ladero JM, et al. Association of CYP2C9 genotypes leading to high enzyme activity and colorectal cancer risk. Carcinogenesis. 2001;22:1323–6. https://doi.org/10.1093/carcin/22.8.1323.
Louet M, Labbe CM, Fagnen C, et al. Insights into molecular mechanisms of drug metabolism dysfunction of human CYP2C9*30. PLoS One. 2018;13:e0197249. https://doi.org/10.1371/journal.pone.0197249.
Tsunedomi R, Iizuka N, Hamamoto Y, et al. Patterns of expression of cytochrome P450 genes in progression of hepatitis C virus-associated hepatocellular carcinoma. Int J Oncol. 2005;27:661–7.
Xu XR, Huang J, Xu ZG, et al. Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci USA. 2001;98:15089–94. https://doi.org/10.1073/pnas.241522398.
Hu DG, Marri S, McKinnon RA, et al. Deregulation of the genes that are involved in drug absorption, distribution, metabolism, and excretion in hepatocellular carcinoma. J Pharmacol Exp Ther. 2019;368:363–81. https://doi.org/10.1124/jpet.118.255018.
Shen J, Wang S, Zhang YJ, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55:1799–808. https://doi.org/10.1002/hep.25569.
Nogueira V, Hay N. Molecular pathways: reactive oxygen species homeostasis in cancer cells and implications for cancer therapy. Clin Cancer Res. 2013;19:4309–14. https://doi.org/10.1158/1078-0432.CCR-12-1424.
Ozaslan MS, Balci N, Demir Y, et al. Inhibition effects of some antidepressant drugs on pentose phosphate pathway enzymes. Environ Toxicol Pharmacol. 2019;72:103244. https://doi.org/10.1016/j.etap.2019.103244.
Caliskan B, Ozturk Kesebir A, Demir Y, et al. The effect of brimonidine and proparacaine on metabolic enzymes: glucose-6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, and glutathione reductase. Biotechnol Appl Biochem. 2021. https://doi.org/10.1002/bab.2107.
Alfarouk KO, Ahmed SBM, Elliott RL, et al. The pentose phosphate pathway dynamics in cancer and ıts dependency on ıntracellular pH. Metabolites. 2020. https://doi.org/10.3390/metabo10070285.
Kowalik MA, Columbano A, Perra A. Emerging role of the pentose phosphate pathway in hepatocellular carcinoma. Front Oncol. 2017;7:87. https://doi.org/10.3389/fonc.2017.00087.
Demir Y, Turkes C, Beydemir S. Molecular docking studies and inhibition properties of some antineoplastic agents against paraoxonase-I. Anticancer Agents Med Chem. 2020;20:887–96. https://doi.org/10.2174/1871520620666200218110645.
Demir Y. The behaviour of some antihypertension drugs on human serum paraoxonase-1: an important protector enzyme against atherosclerosis. J Pharm Pharmacol. 2019;71:1576–83. https://doi.org/10.1111/jphp.13144.
Demir Y. Naphthoquinones, benzoquinones, and anthraquinones: molecular docking, ADME and inhibition studies on human serum paraoxonase-1 associated with cardiovascular diseases. Drug Dev Res. 2020;81:628–36. https://doi.org/10.1002/ddr.21667.
Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39:347–54. https://doi.org/10.1016/j.tibs.2014.06.005.
Gonul Baltaci N, Guler C, Ceylan H, et al. In vitro and in vivo effects of iron on the expression and activity of glucose 6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, and glutathione reductase in rat spleen. J Biochem Mol Toxicol. 2018. https://doi.org/10.1002/jbt.22229.
Kocpinar EF, Gonul Baltaci N, Ceylan H, et al. Effect of a prolonged dietary iron intake on the gene expression and activity of the testicular antioxidant defense system in rats. Biol Trace Elem Res. 2020;195:135–41. https://doi.org/10.1007/s12011-019-01817-0.
Ma X, Wang L, Huang D, et al. Polo-like kinase 1 coordinates biosynthesis during cell cycle progression by directly activating pentose phosphate pathway. Nat Commun. 2017;8:1506. https://doi.org/10.1038/s41467-017-01647-5.
Ye H, Huang H, Cao F, et al. HSPB1 enhances SIRT2-mediated G6PD activation and promotes glioma cell proliferation. PLoS One. 2016;11:e0164285. https://doi.org/10.1371/journal.pone.0164285.
Debeb BG, Lacerda L, Larson R, et al. Histone deacetylase inhibitor-induced cancer stem cells exhibit high pentose phosphate pathway metabolism. Oncotarget. 2016;7:28329–39. https://doi.org/10.18632/oncotarget.8631.
Ham M, Lee JW, Choi AH, et al. Macrophage glucose-6-phosphate dehydrogenase stimulates proinflammatory responses with oxidative stress. Mol Cell Biol. 2013;33:2425–35. https://doi.org/10.1128/MCB.01260-12.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of ınterest
The author declares that there is no conflict of interest with any financial organization or corporation or individual that can inappropriately influence this work.
Ethical approval
This study does not contain any studies with human participants or animals performed by the author.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ceylan, H. Identification of hub genes associated with obesity-induced hepatocellular carcinoma risk based on integrated bioinformatics analysis. Med Oncol 38, 63 (2021). https://doi.org/10.1007/s12032-021-01510-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-021-01510-0